Page 76 - Genetics_From_Genes_to_Genomes_6th_FULL_Part3
P. 76
370 Chapter 11 Analyzing Genomic Variation
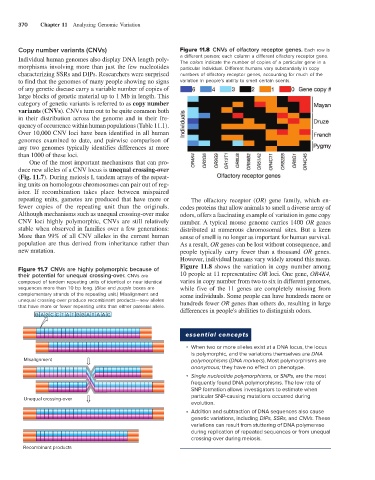
Copy number variants (CNVs) Figure 11.8 CNVs of olfactory receptor genes. Each row is
Individual human genomes also display DNA length poly- a different person; each column a different olfactory receptor gene.
The colors indicate the number of copies of a particular gene in a
morphisms involving more than just the few nucleotides particular individual. Different humans vary substantially in copy
characterizing SSRs and DIPs. Researchers were surprised numbers of olfactory receptor genes, accounting for much of the
to find that the genomes of many people showing no signs variation in people’s ability to smell certain scents.
of any genetic disease carry a variable number of copies of
large blocks of genetic material up to 1 Mb in length. This
category of genetic variants is referred to as copy number
variants (CNVs). CNVs turn out to be quite common both
in their distribution across the genome and in their fre-
quency of occurrence within human populations (Table 11.1).
Over 10,000 CNV loci have been identified in all human
genomes examined to date, and pairwise comparison of
any two genomes typically identifies differences at more
than 1000 of these loci.
One of the most important mechanisms that can pro-
duce new alleles of a CNV locus is unequal crossing-over
(Fig. 11.7). During meiosis I, tandem arrays of the repeat-
ing units on homologous chromosomes can pair out of reg-
ister. If recombination takes place between mispaired
repeating units, gametes are produced that have more or The olfactory receptor (OR) gene family, which en-
fewer copies of the repeating unit than the originals. codes proteins that allow animals to smell a diverse array of
Although mechanisms such as unequal crossing-over make odors, offers a fascinating example of variation in gene copy
CNV loci highly polymorphic, CNVs are still relatively number. A typical mouse genome carries 1400 OR genes
stable when observed in families over a few generations: distributed at numerous chromosomal sites. But a keen
More than 99% of all CNV alleles in the current human sense of smell is no longer as important for human survival.
population are thus derived from inheritance rather than As a result, OR genes can be lost without consequence, and
new mutation. people typically carry fewer than a thousand OR genes.
However, individual humans vary widely around this mean.
Figure 11.8 shows the variation in copy number among
Figure 11.7 CNVs are highly polymorphic because of
their potential for unequal crossing-over. CNVs are 10 people at 11 representative OR loci. One gene, OR4K4,
composed of tandem repeating units of identical or near identical varies in copy number from two to six in different genomes,
sequences more than 10 bp long. (Blue and purple boxes are while five of the 11 genes are completely missing from
complementary strands of the repeating unit.) Misalignment and some individuals. Some people can have hundreds more or
unequal crossing-over produce recombinant products—new alleles hundreds fewer OR genes than others do, resulting in large
that have more or fewer repeating units than either parental allele.
differences in people's abilities to distinguish odors.
G A G C C T A T G G A T A A C
essential concepts
• When two or more alleles exist at a DNA locus, the locus
is polymorphic, and the variations themselves are DNA
Misalignment polymorphisms (DNA markers). Most polymorphisms are
anonymous; they have no effect on phenotype.
• Single nucleotide polymorphisms, or SNPs, are the most
frequently found DNA polymorphisms. The low rate of
SNP formation allows investigators to estimate when
particular SNP-causing mutations occurred during
Unequal crossing-over
evolution.
• Addition and subtraction of DNA sequences also cause
genetic variations, including DIPs, SSRs, and CNVs. These
variations can result from stuttering of DNA polymerase
during replication of repeated sequences or from unequal
crossing-over during meiosis.
Recombinant products