Page 72 - Genetics_From_Genes_to_Genomes_6th_FULL_Part3
P. 72
366 Chapter 11 Analyzing Genomic Variation
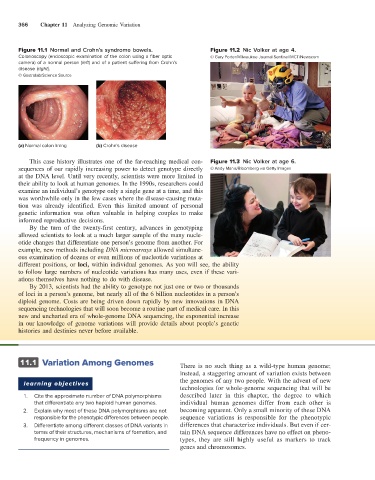
Figure 11.1 Normal and Crohn’s syndrome bowels. Figure 11.2 Nic Volker at age 4.
Colonoscopy (endoscopic examination of the colon using a fiber optic © Gary Porter/Milwaukee Journal Sentinel/MCT/Newscom
camera) of a normal person (left) and of a patient suffering from Crohn’s
disease (right).
© Gastrolab/Science Source
(a) Normal colon lining (b) Crohn’s disease
This case history illustrates one of the far-reaching medical con- Figure 11.3 Nic Volker at age 6.
sequences of our rapidly increasing power to detect genotype directly © Andy Manis/Bloomberg via Getty Images
at the DNA level. Until very recently, scientists were more limited in
their ability to look at human genomes. In the 1990s, researchers could
examine an individual’s genotype only a single gene at a time, and this
was worthwhile only in the few cases where the disease-causing muta-
tion was already identified. Even this limited amount of personal
genetic information was often valuable in helping couples to make
informed reproductive decisions.
By the turn of the twenty-first century, advances in genotyping
allowed scientists to look at a much larger sample of the many nucle-
otide changes that differentiate one person’s genome from another. For
example, new methods including DNA microarrays allowed simultane-
ous examination of dozens or even millions of nucleotide variations at
different positions, or loci, within individual genomes. As you will see, the ability
to follow large numbers of nucleotide variations has many uses, even if these vari-
ations themselves have nothing to do with disease.
By 2013, scientists had the ability to genotype not just one or two or thousands
of loci in a person’s genome, but nearly all of the 6 billion nucleotides in a person’s
diploid genome. Costs are being driven down rapidly by new innovations in DNA
sequencing technologies that will soon become a routine part of medical care. In this
new and uncharted era of whole-genome DNA sequencing, the exponential increase
in our knowledge of genome variations will provide details about people’s genetic
histories and destinies never before available.
11.1 Variation Among Genomes There is no such thing as a wild-type human genome;
instead, a staggering amount of variation exists between
the genomes of any two people. With the advent of new
learning objectives
technologies for whole-genome sequencing that will be
1. Cite the approximate number of DNA polymorphisms described later in this chapter, the degree to which
that differentiate any two haploid human genomes. individual human genomes differ from each other is
2. Explain why most of these DNA polymorphisms are not becoming apparent. Only a small minority of these DNA
responsible for the phenotypic differences between people. sequence variations is responsible for the phenotypic
3. Differentiate among different classes of DNA variants in differences that characterize individuals. But even if cer-
terms of their structures, mechanisms of formation, and tain DNA sequence differences have no effect on pheno-
frequency in genomes. types, they are still highly useful as markers to track
genes and chromosomes.