Page 74 - Genetics_From_Genes_to_Genomes_6th_FULL_Part3
P. 74
368 Chapter 11 Analyzing Genomic Variation
polymorphisms in both men’s genomes, and indeed in all several of the many mechanisms that can give rise to SNPs,
genomes, are located either between genes or in introns, including rare mistakes in DNA replication, or exposure of
consistent with the idea that most DNA changes do not the genome to mutagenic chemicals or radiation in the
alter phenotype. environment.
The existence of such a vast number of anonymous DNA Despite the existence of so many types of events that
polymorphisms that distinguish different human genomes can create SNPs, the per-base spontaneous mutation rate is
presents both challenges and opportunities for geneticists. still less than one in 30 million (by some estimates as low
The challenges are clear: How can we sort out the millions of as one in 100 million) per generation. This number is so
polymorphisms in anyone’s DNA to find the few that are low that most SNPs are biallelic in human populations,
relevant to traits such as genetic diseases? The opportunities with only two of the four possible nucleotide pairs repre-
lie in the fact that even if a polymorphism is anonymous, with sented. The low mutation rate of SNPs allows researchers
no effect on phenotype, it still serves as a signpost in the to trace back each individual SNP to a genomic change that
genome, a DNA marker. You will see that researchers can occurred once in a single ancestral genome. The low muta-
use anonymous polymorphic loci to help locate the nearby tion frequency also means that those people who did not
mutations that are actually responsible for inherited diseases. inherit this changed nucleotide (called the derived allele)
have a more ancient ancestral allele that was probably
present long before the human species took form.
Genetic Variants Occur in Several Types If a SNP exists, geneticists can take advantage of the
close relationship between the human and chimpanzee
Geneticists usually place polymorphic DNA loci into one genomes to determine which allele is ancient and which
of the four categories shown in Table 11.1 based on the is the derived allele resulting from a relatively recent mu-
number and kinds of nucleotide pairs involved. Although tational event. Figure 11.6 compares a small region of
the borders between these classes are fuzzy and overlap to
some extent, the categories help researchers describe what
a particular genetic variant looks like. A useful generaliza-
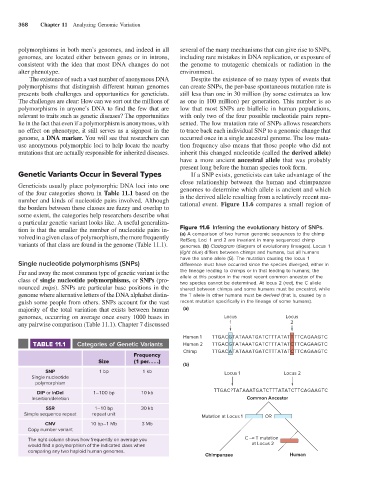
tion is that the smaller the number of nucleotide pairs in- Figure 11.6 Inferring the evolutionary history of SNPs.
volved in a given class of polymorphism, the more frequently (a) A comparison of two human genomic sequences to the chimp
RefSeq. Loci 1 and 2 are invariant in many sequenced chimp
variants of that class are found in the genome (Table 11.1). genomes. (b) Cladogram (diagram of evolutionary lineages). Locus 1
(light blue) differs between chimps and humans, but all humans
have the same allele (G). The mutation causing the locus 1
Single nucleotide polymorphisms (SNPs) difference must have occurred since the species diverged, either in
Far and away the most common type of genetic variant is the the lineage leading to chimps or in that leading to humans; the
class of single nucleotide polymorphisms, or SNPs (pro- allele at this position in the most recent common ancestor of the
two species cannot be determined. At locus 2 (red), the C allele
nounced snips). SNPs are particular base positions in the shared between chimps and some humans must be ancestral, while
genome where alternative letters of the DNA alphabet distin- the T allele in other humans must be derived (that is, caused by a
guish some people from others. SNPs account for the vast recent mutation specifically in the lineage of some humans).
majority of the total variation that exists between human (a)
genomes, occurring on average once every 1000 bases in Locus Locus
any pairwise comparison (Table 11.1). Chapter 7 discussed 1 2
Human 1 TTGACGTATAAATGATCTTTATATT TTCAGAAGTC
TABLE 11.1 Categories of Genetic Variants Human 2 TTGACGTATAAATGATCTTTATATCTTCAGAAGTC
Chimp TTGACATATAAATGATCTTTATATCTTCAGAAGTC
Frequency
Size (1 per. . . .)
(b)
SNP 1 bp 1 kb Locus 1 Locus 2
Single nucleotide
polymorphism
TTGAC?TATAAATGATCTTTATATCTTCAGAAGTC
DIP or InDel 1–100 bp 10 kb
Insertion/deletion Common Ancestor
SSR 1–10 bp 30 kb
Simple sequence repeat repeat unit Mutation at Locus 1 OR
CNV 10 bp–1 Mb 3 Mb
Copy number variant
The right column shows how frequently on average you C T mutation
would find a polymorphism of the indicated class when at Locus 2
comparing any two haploid human genomes.
Chimpanzee Human