Page 73 - Genetics_From_Genes_to_Genomes_6th_FULL_Part3
P. 73
11.1 Variation Among Genomes 367
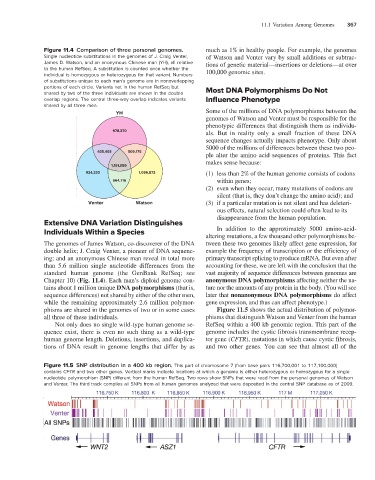
Figure 11.4 Comparison of three personal genomes. much as 1% in healthy people. For example, the genomes
Single nucleotide substitutions in the genomes of J. Craig Venter, of Watson and Venter vary by small additions or subtrac-
James D. Watson, and an anonymous Chinese man (YH), all relative tions of genetic material—insertions or deletions—at over
to the human RefSeq. A substitution is counted once whether the 100,000 genomic sites.
individual is homozygous or heterozygous for that variant. Numbers
of substitutions unique to each man’s genome are in nonoverlapping
portions of each circle. Variants not in the human RefSeq but Most DNA Polymorphisms Do Not
shared by two of the three individuals are shown in the double
overlap regions. The central three-way overlap indicates variants Influence Phenotype
shared by all three men.
YH Some of the millions of DNA polymorphisms between the
genomes of Watson and Venter must be responsible for the
phenotypic differences that distinguish them as individu-
978,370 als. But in reality only a small fraction of these DNA
sequence changes actually impacts phenotype. Only about
5000 of the millions of differences between these two peo-
435,493 509,175
ple alter the amino acid sequences of proteins. This fact
makes sense because:
1,151,059
924,333 1,096,873 (1) less than 2% of the human genome consists of codons
564,716 within genes;
(2) even when they occur, many mutations of codons are
silent (that is, they don’t change the amino acid); and
Venter Watson (3) if a particular mutation is not silent and has deleteri-
ous effects, natural selection could often lead to its
disappearance from the human population.
Extensive DNA Variation Distinguishes
In addition to the approximately 5000 amino-acid-
Individuals Within a Species altering mutations, a few thousand other polymorphisms be-
The genomes of James Watson, co-discoverer of the DNA tween these two genomes likely affect gene expression, for
double helix; J. Craig Venter, a pioneer of DNA sequenc- example the frequency of transcription or the efficiency of
ing; and an anonymous Chinese man reveal in total more primary transcript splicing to produce mRNA. But even after
than 5.6 million single nucleotide differences from the accounting for these, we are left with the conclusion that the
standard human genome (the GenBank RefSeq; see vast majority of sequence differences between genomes are
Chapter 10) (Fig. 11.4). Each man’s diploid genome con- anonymous DNA polymorphisms affecting neither the na-
tains about 1 million unique DNA polymorphisms (that is, ture nor the amounts of any protein in the body. (You will see
sequence differences) not shared by either of the other men, later that nonanonymous DNA polymorphisms do affect
while the remaining approximately 2.6 million polymor- gene expression, and thus can affect phenotype.)
phisms are shared in the genomes of two or in some cases Figure 11.5 shows the actual distribution of polymor-
all three of these individuals. phisms that distinguish Watson and Venter from the human
Not only does no single wild-type human genome se- RefSeq within a 400 kb genomic region. This part of the
quence exist, there is even no such thing as a wild-type genome includes the cystic fibrosis transmembrane recep-
human genome length. Deletions, insertions, and duplica- tor gene (CFTR), mutations in which cause cystic fibrosis,
tions of DNA result in genome lengths that differ by as and two other genes. You can see that almost all of the
Figure 11.5 SNP distribution in a 400 kb region. This part of chromosome 7 (from base pairs 116,700,001 to 117,100,000)
contains CFTR and two other genes. Vertical marks indicate locations at which a genome is either heterozygous or homozygous for a single
nucleotide polymorphism (SNP) different from the human RefSeq. Two rows show SNPs that were read from the personal genomes of Watson
and Venter. The third track compiles all SNPs from all human genomes analyzed that were deposited in the central SNP database as of 2009.