Page 52 - Genetics_From_Genes_to_Genomes_6th_FULL_Part2
P. 52
6.6 Site-Specific Recombination 211
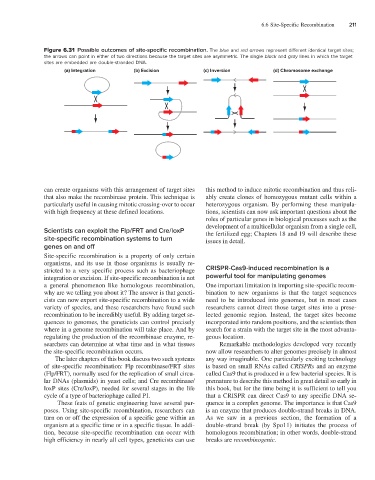
Figure 6.31 Possible outcomes of site-specific recombination. The blue and red arrows represent different identical target sites;
the arrows can point in either of two directions because the target sites are asymmetric. The single black and gray lines in which the target
sites are embedded are double-stranded DNA.
(a) Integration (b) Excision (c) Inversion (d) Chromosome exchange
can create organisms with this arrangement of target sites this method to induce mitotic recombination and thus reli-
that also make the recombinase protein. This technique is ably create clones of homozygous mutant cells within a
particularly useful in causing mitotic crossing-over to occur heterozygous organism. By performing these manipula-
with high frequency at these defined locations. tions, scientists can now ask important questions about the
roles of particular genes in biological processes such as the
development of a multicellular organism from a single cell,
Scientists can exploit the Flp/FRT and Cre/loxP the fertilized egg; Chapters 18 and 19 will describe these
site-specific recombination systems to turn issues in detail.
genes on and off
Site-specific recombination is a property of only certain
organisms, and its use in those organisms is usually re-
stricted to a very specific process such as bacteriophage CRISPR-Cas9-induced recombination is a
integration or excision. If site-specific recombination is not powerful tool for manipulating genomes
a general phenomenon like homologous recombination, One important limitation in importing site-specific recom-
why are we telling you about it? The answer is that geneti- bination to new organisms is that the target sequences
cists can now export site-specific recombination to a wide need to be introduced into genomes, but in most cases
variety of species, and these researchers have found such researchers cannot direct those target sites into a prese-
recombination to be incredibly useful. By adding target se- lected genomic region. Instead, the target sites become
quences to genomes, the geneticists can control precisely incorporated into random positions, and the scientists then
where in a genome recombination will take place. And by search for a strain with the target site in the most advanta-
regulating the production of the recombinase enzyme, re- geous location.
searchers can determine at what time and in what tissues Remarkable methodologies developed very recently
the site-specific recombination occurs. now allow researchers to alter genomes precisely in almost
The later chapters of this book discuss two such systems any way imaginable. One particularly exciting technology
of site-specific recombination: Flp recombinase/FRT sites is based on small RNAs called CRISPRs and an enzyme
(Flp/FRT), normally used for the replication of small circu- called Cas9 that is produced in a few bacterial species. It is
lar DNAs (plasmids) in yeast cells; and Cre recombinase/ premature to describe this method in great detail so early in
loxP sites (Cre/loxP), needed for several stages in the life this book, but for the time being it is sufficient to tell you
cycle of a type of bacteriophage called P1. that a CRISPR can direct Cas9 to any specific DNA se-
These feats of genetic engineering have several pur- quence in a complex genome. The importance is that Cas9
poses. Using site-specific recombination, researchers can is an enzyme that produces double-strand breaks in DNA.
turn on or off the expression of a specific gene within an As we saw in a previous section, the formation of a
organism at a specific time or in a specific tissue. In addi- double-strand break (by Spo11) initiates the process of
tion, because site-specific recombination can occur with homologous recombination; in other words, double-strand
high efficiency in nearly all cell types, geneticists can use breaks are recombinogenic.