Page 51 - Genetics_From_Genes_to_Genomes_6th_FULL_Part2
P. 51
210 Chapter 6 DNA Structure, Replication, and Recombination
DNA sequences. Site-specific recombination is crossing- between two target sites in a single chromosome is removed
over that occurs only between two specific DNA target to create two independent DNA molecules (Fig. 6.31b). If
sites that are usually less than 200 base pairs long. Site- a bacteriophage genome was previously integrated into the
specific recombination is much simpler at the molecular host chromosome, excision is crucial to allow the bacterio-
level than is the homologous recombination discussed in phage genome to extricate itself and then to become incor-
the previous section. In particular, in most systems of porated in the virus particle.
site-specific recombination, a single protein logically A third potential outcome of site-specific recombina-
called a recombinase is sufficient to catalyze all the tion systems is the inversion of a segment of DNA that is
breakage and joining steps of the process. If you are curi- located between the two target sites (Fig. 6.31c). As you
ous, Fig. 6.30 depicts the mode of action of one class of can imagine, such inversion could constitute a molecular
such recombinases. switch between two configurations of the same chromo-
The organisms that take advantage of site-specific some. The in-between segment is oriented in one direction
recombination include certain kinds of bacteriophages in one state and in the other direction in the other state.
that use this process for the integration (incorporation) A final mode of site-specific recombination can occur
of their small, circular genome into the chromosome of if the target site is found at the same position on each of two
the host bacterium (Fig. 6.31a). In this way, the bacteri- homologous chromosomes. Action of the recombinase on
ophage DNA “hitchhikes” along with the bacterial chro- these target sites will result in the reshuffling of regions on
mosome: When the host DNA replicates, so does the nonsister chromatids, an outcome that leads to recombinant
integrated bacteriophage genome. chromosomes (Fig. 6.31d). To our knowledge, this situa-
Site-specific recombination is also important for the tion is not normally encountered in organisms that natu-
reverse process of excision, in which the DNA integrated rally use site-specific recombination. However, geneticists
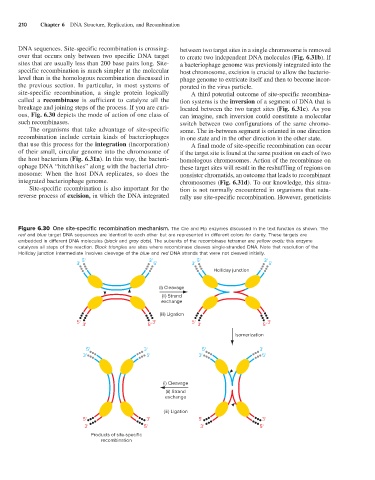
Figure 6.30 One site-specific recombination mechanism. The Cre and Flp enzymes discussed in the text function as shown. The
red and blue target DNA sequences are identical to each other but are represented in different colors for clarity. These targets are
embedded in different DNA molecules (black and gray dots). The subunits of the recombinase tetramer are yellow ovals; this enzyme
catalyzes all steps of the reaction. Black triangles are sites where recombinase cleaves single-stranded DNA. Note that resolution of the
Holliday junction intermediate involves cleavage of the blue and red DNA strands that were not cleaved initially.
5' 3' 5' 3'
3' 5' 3' 5'
Holliday junction
(i) Cleavage
(ii) Strand
exchange
(iii) Ligation
5' 3' 5' 3' 5' 3' 5' 3'
Isomerization
5' 3' 5' 3'
3' 5' 3' 5'
(i) Cleavage
(ii) Strand
exchange
(iii) Ligation
5' 3' 5' 3'
3' 5' 3' 5'
Products of site-specific
recombination