Page 137 - Genetics_From_Genes_to_Genomes_6th_FULL_Part2
P. 137
296 Chapter 8 Gene Expression: The Flow of Information from DNA to RNA to Protein
you examine the figure, note the following points about the important roles played by protein translation factors, which
flow of information during translation: help shepherd mRNAs and tRNAs to their proper locations
on the ribosome. Some translation factors also carry GTP
∙ The first codon to be translated—the initiation codon— to the ribosome. Hydrolysis of the high-energy bonds in the
is an AUG set in a special context at the 5′ end of the GTP helps power certain molecular movements, including
gene’s reading frame (never precisely at the 5′ end of translocation of the ribosome along the mRNA.
the mRNA).
∙ Special initiating tRNAs carrying a modified form of
methionine called formylmethionine ( fMet) recognize Polypeptides Can Be Modified
the initiation codon. After Translation
∙ The ribosome moves along the mRNA in the 5′-to-3′ di- Protein structure is not irrevocably fixed at the completion
rection, revealing successive codons in a stepwise fashion.
∙ At each step of translation, the polypeptide grows by of translation. Several different processes may subse-
quently modify a polypeptide’s structure. Cleavage may
the addition to its C terminus of the next amino acid remove amino acids, such as the N-terminal fMet, from a
in the chain.
∙ Translation terminates when the ribosome reaches a polypeptide, or it may generate several smaller polypep-
tides from one larger product of translation (Fig. 8.26a). In
UAA, UAG, or UGA nonsense codon at the 3′ end of the latter case, the larger polypeptide made before it is cleaved
the gene’s reading frame.
These points explain the biochemical basis of colin- into smaller polypeptides is often called a polyprotein. Also,
some proteins are synthesized in inactive forms called
earity, that is, the correspondence between the 5′-to-3′ zymogens that are activated by enzymatic cleavage that
direction in the mRNA and the N-terminus-to- removes an N-terminal prosegment.
C-terminus direction in the resulting polypeptide.
Enzymatic addition of chemical constituents, such as
During elongation, the translation machinery adds phosphate groups, carbohydrates, fatty acids, or even other
about 2–15 amino acids per second to the growing chain. small peptides to specific amino acids may also modify a pol-
The speed is higher in prokaryotes and lower in eukaryotes. ypeptide after translation (Fig. 8.26b). Such changes to poly-
At these rates, construction of an average-size 300-amino- peptides are known as posttranslational modifications.
acid polypeptide (from an average-length mRNA that is Posttranslational changes to a protein can be very important:
about 1000 nucleotides) could take as little as 20 seconds For example, the biochemical function of many enzymes de-
or as long as 2.5 minutes. pends directly on the addition (or sometimes removal) of
Several details have been left out of Fig. 8.25 so that phosphate groups. Posttranslational modifications may alter
you can concentrate on the flow of information during the way a protein folds, its ability to interact with other pro-
translation. In particular, this figure does not depict the teins, its stability, its activity, or its location in the cell.
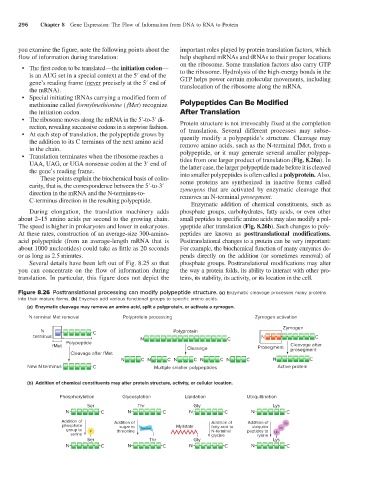
Figure 8.26 Posttranslational processing can modify polypeptide structure. (a) Enzymatic cleavage processes many proteins
into their mature forms. (b) Enyzmes add various functional groups to specific amino acids.
(a) Enzymatic cleavage may remove an amino acid, split a polyprotein, or activate a zymogen.
N-terminal Met removal Polyprotein processing Zymogen activation
Zymogen
N C Polyprotein
terminus N C N C
Polypeptide
fMet Cleavage after
Cleavage Prosegment prosegment
Cleavage after fMet
N C N C N C N C N C N C
New N terminus C Multiple smaller polypeptides Active protein
(b) Addition of chemical constituents may alter protein structure, activity, or cellular location.
Phosphorylation Glycosylation Lipidation Ubiquitination
Ser Thr Gly Lys
N C N C N C N C
Addition of Addition of Addition of Addition of Ub
phosphate sugar to Myristate fatty acid to ubiquitin
group to P threonine hexose N-terminal peptides to Ub
serine glycine lysine Ub
Ser Thr Gly Lys
N C N C N C N C