Page 129 - Genetics_From_Genes_to_Genomes_6th_FULL_Part2
P. 129
288 Chapter 8 Gene Expression: The Flow of Information from DNA to RNA to Protein
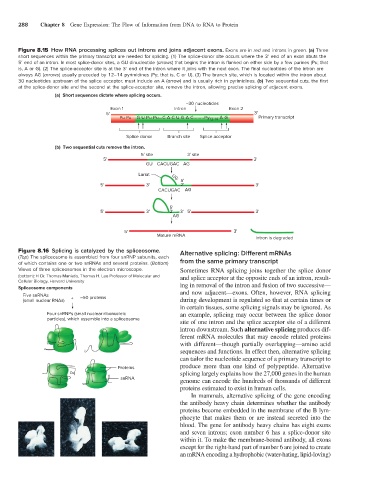
Figure 8.15 How RNA processing splices out introns and joins adjacent exons. Exons are in red and introns in green. (a) Three
short sequences within the primary transcript are needed for splicing. (1) The splice-donor site occurs where the 3′ end of an exon abuts the
5′ end of an intron. In most splice-donor sites, a GU dinucleotide (arrows) that begins the intron is flanked on either side by a few purines (Pu; that
is, A or G). (2) The splice-acceptor site is at the 3′ end of the intron where it joins with the next exon. The final nucleotides of the intron are
always AG (arrows) usually preceded by 12–14 pyrimidines (Py; that is, C or U). (3) The branch site, which is located within the intron about
30 nucleotides upstream of the splice acceptor, must include an A (arrow) and is usually rich in pyrimidines. (b) Two sequential cuts, the first
at the splice-donor site and the second at the splice-acceptor site, remove the intron, allowing precise splicing of adjacent exons.
(a) Short sequences dictate where splicing occurs.
~30 nucleotides
Exon 1 Intron Exon 2
5' 3'
Pu Pu G U Pu Pu...C A C U G A C........Py 12-14 A G Primary transcript
Splice donor Branch site Splice acceptor
(b) Two sequential cuts remove the intron.
5' site 3' site
5' 3'
GU CACUGAC AG
Lariat UG
5'
5' 3' 2' 3'
CACUGAC AG
5'
5' 3' 2' 3' 5' 3'
AG
5' 3'
Mature mRNA
Intron is degraded
Figure 8.16 Splicing is catalyzed by the spliceosome. Alternative splicing: Different mRNAs
(Top) The spliceosome is assembled from four snRNP subunits, each
of which contains one or two snRNAs and several proteins. (Bottom) from the same primary transcript
Views of three spliceosomes in the electron microscope. Sometimes RNA splicing joins together the splice donor
(bottom): © Dr. Thomas Maniatis, Thomas H. Lee Professor of Molecular and and splice acceptor at the opposite ends of an intron, result
Cellular Biology, Harvard University
Spliceosome components ing in removal of the intron and fusion of two successive—
Five snRNAs 50 proteins and now adjacent—exons. Often, however, RNA splicing
(small nuclear RNAs) + ~ during development is regulated so that at certain times or
in certain tissues, some splicing signals may be ignored. As
Four snRNPs (small nuclear ribonucleic an example, splicing may occur between the splice donor
particles), which assemble into a spliceosome
site of one intron and the splice acceptor site of a different
intron downstream. Such alternative splicing produces dif
ferent mRNA molecules that may encode related proteins
with different—though partially overlapping—amino acid
sequences and functions. In effect then, alternative splicing
can tailor the nucleotide sequence of a primary transcript to
Proteins produce more than one kind of polypeptide. Alternative
splicing largely explains how the 27,000 genes in the human
snRNA
genome can encode the hundreds of thousands of different
proteins estimated to exist in human cells.
In mammals, alternative splicing of the gene encoding
the antibody heavy chain determines whether the antibody
proteins become embedded in the membrane of the B lym
phocyte that makes them or are instead secreted into the
blood. The gene for antibody heavy chains has eight exons
and seven introns; exon number 6 has a splicedonor site
within it. To make the membranebound antibody, all exons
except for the righthand part of number 6 are joined to create
an mRNA encoding a hydrophobic (waterhating, lipidloving)