Page 128 - Genetics_From_Genes_to_Genomes_6th_FULL_Part2
P. 128
8.2 Transcription: From DNA to RNA 287
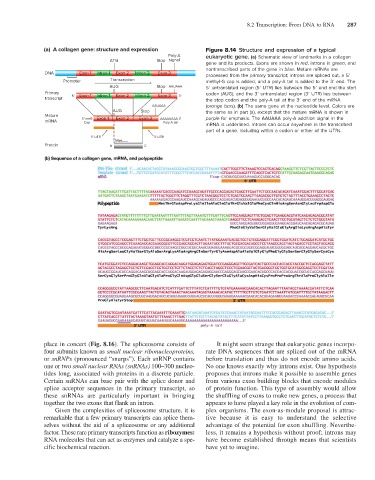
(a) A collagen gene: structure and expression Figure 8.14 Structure and expression of a typical
Poly-A eukaryotic gene. (a) Schematic view of landmarks in a collagen
ATG Stop signal
gene and its products. Exons are shown in red, introns in green, and
nontranscribed parts of the gene in blue. Mature mRNAs are
DNA Exon 1 Intron 1 Exon 2 Intron 2 Exon 3 processed from the primary transcript; introns are spliced out, a 5′
Promoter Transcription methyl-G cap is added, and a poly-A tail is added to the 3′ end. The
AUG Stop AAUAAA 5′ untranslated region (5′ UTR) lies between the 5′ end and the start
Primary 5' Exon 1 Intron 1 Exon 2 Intron 2 Exon 3 3' codon (AUG), and the 3′ untranslated region (3′ UTR) lies between
transcript the stop codon and the poly-A tail at the 3′ end of the mRNA
AAUAAA (orange bars). (b) The same gene at the nucleotide level. Colors are
AUG Stop the same as in part (a), except that the mature mRNA is shown in
Mature 5' meG Exon 1 Exon 2 Exon 3 AAAAAAAA 3' purple for emphasis. The AAUAAA poly-A addition signal in the
mRNA Cap Poly-A tail
mRNA is underlined. Introns can occur anywhere in the transcribed
part of a gene, including within a codon or either of the UTRs.
5' UTR 3' UTR
Met......
Protein N C
place in concert (Fig. 8.16). The spliceosome consists of It might seem strange that eukaryotic genes incorpo
four subunits known as small nuclear ribonucleoproteins, rate DNA sequences that are spliced out of the mRNA
or snRNPs (pronounced “snurps”). Each snRNP contains before translation and thus do not encode amino acids.
one or two small nuclear RNAs (snRNAs) 100–300 nucleo No one knows exactly why introns exist. One hypothesis
tides long, associated with proteins in a discrete particle. proposes that introns make it possible to assemble genes
Certain snRNAs can base pair with the splice donor and from various exon building blocks that encode modules
splice acceptor sequences in the primary transcript, so of protein function. This type of assembly would allow
these snRNAs are particularly important in bringing the shuffling of exons to make new genes, a process that
together the two exons that flank an intron. appears to have played a key role in the evolution of com
Given the complexities of spliceosome structure, it is plex organisms. The exonasmodule proposal is attrac
remarkable that a few primary transcripts can splice them tive because it is easy to understand the selective
selves without the aid of a spliceosome or any additional advantage of the potential for exon shuffling. Neverthe
factor. These rare primary transcripts function as ribozymes: less, it remains a hypothesis without proof; introns may
RNA molecules that can act as enzymes and catalyze a spe have become established through means that scientists
cific biochemical reaction. have yet to imagine.