Page 127 - Genetics_From_Genes_to_Genomes_6th_FULL_Part2
P. 127
286 Chapter 8 Gene Expression: The Flow of Information from DNA to RNA to Protein
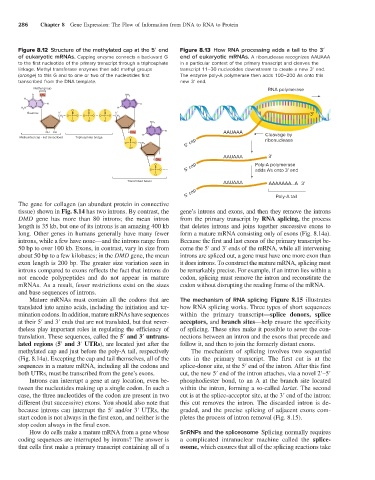
Figure 8.12 Structure of the methylated cap at the 5′ end Figure 8.13 How RNA processing adds a tail to the 3′
of eukaryotic mRNAs. Capping enzyme connects a backward G end of eukaryotic mRNAs. A ribonuclease recognizes AAUAAA
to the first nucleotide of the primary transcript through a triphosphate in a particular context of the primary transcript and cleaves the
linkage. Methyl transferase enzymes then add methyl groups transcript 11–30 nucleotides downstream to create a new 3′ end.
(orange) to this G and to one or two of the nucleotides first The enzyme poly-A polymerase then adds 100–200 As onto this
transcribed from the DNA template. new 3′ end.
Methyl group RNA polymerase
O CH 3 NH 2
H C N C N
N C N C
C C C H C C C H
H 2 N N N H N N
O O O
Guanine 5' 5' 3'
CH 2 O P O O P O O P O CH 2
O – O – O –
NH 2
C N
N C
OH OH OCH 3 C C C H AAUAAA
H N N Cleavage by
Methylated cap - not transcribed Triphosphate bridge O
O P O CH 2 5' cap ribonuclease
O –
AAUAAA 3'
OCH 3
O 5' cap Poly-A polymerase
O P O .... adds A's onto 3' end
O –
Transcribed bases AAUAAA AAAAAAA...A 3'
5' cap Poly-A tail
The gene for collagen (an abundant protein in connective
tissue) shown in Fig. 8.14 has two introns. By contrast, the gene’s introns and exons, and then they remove the introns
DMD gene has more than 80 introns; the mean intron from the primary transcript by RNA splicing, the process
length is 35 kb, but one of its introns is an amazing 400 kb that deletes introns and joins together successive exons to
long. Other genes in humans generally have many fewer form a mature mRNA consisting only of exons (Fig. 8.14a).
introns, while a few have none—and the introns range from Because the first and last exons of the primary transcript be
50 bp to over 100 kb. Exons, in contrast, vary in size from come the 5′ and 3′ ends of the mRNA, while all intervening
about 50 bp to a few kilobases; in the DMD gene, the mean introns are spliced out, a gene must have one more exon than
exon length is 200 bp. The greater size variation seen in it does introns. To construct the mature mRNA, splicing must
introns compared to exons reflects the fact that introns do be remarkably precise. For example, if an intron lies within a
not encode polypeptides and do not appear in mature codon, splicing must remove the intron and reconstitute the
mRNAs. As a result, fewer restrictions exist on the sizes codon without disrupting the reading frame of the mRNA.
and base sequences of introns.
Mature mRNAs must contain all the codons that are The mechanism of RNA splicing Figure 8.15 illustrates
translated into amino acids, including the initiation and ter how RNA splicing works. Three types of short sequences
mination codons. In addition, mature mRNAs have sequences within the primary transcript—splice donors, splice
at their 5′ and 3′ ends that are not translated, but that never acceptors, and branch sites—help ensure the specificity
theless play important roles in regulating the efficiency of of splicing. These sites make it possible to sever the con
translation. These sequences, called the 5′ and 3′ untrans- nections between an intron and the exons that precede and
lated regions (5′ and 3′ UTRs), are located just after the follow it, and then to join the formerly distant exons.
methylated cap and just before the polyA tail, respectively The mechanism of splicing involves two sequential
(Fig. 8.14a). Excepting the cap and tail themselves, all of the cuts in the primary transcript. The first cut is at the
sequences in a mature mRNA, including all the codons and splicedonor site, at the 5′ end of the intron. After this first
both UTRs, must be transcribed from the gene’s exons. cut, the new 5′ end of the intron attaches, via a novel 2′–5′
Introns can interrupt a gene at any location, even be phosphodiester bond, to an A at the branch site located
tween the nucleotides making up a single codon. In such a within the intron, forming a socalled lariat. The second
case, the three nucleotides of the codon are present in two cut is at the spliceacceptor site, at the 3′ end of the intron;
different (but successive) exons. You should also note that this cut removes the intron. The discarded intron is de
because introns can interrupt the 5′ and/or 3′ UTRs, the graded, and the precise splicing of adjacent exons com
start codon is not always in the first exon, and neither is the pletes the process of intron removal (Fig. 8.15).
stop codon always in the final exon.
How do cells make a mature mRNA from a gene whose SnRNPs and the spliceosome Splicing normally requires
coding sequences are interrupted by introns? The answer is a complicated intranuclear machine called the splice-
that cells first make a primary transcript containing all of a osome, which ensures that all of the splicing reactions take