Page 86 - Genetics_From_Genes_to_Genomes_6th_FULL_Part3
P. 86
380 Chapter 11 Analyzing Genomic Variation
detecting SNP alleles at more than 4 million loci Linkage Analysis with DNA Markers Gives
(Fig. 11.17c). At the time of this writing (2016), the cost of Disease Genes an Approximate
analyzing a sample of genomic DNA is only a few hundred
dollars, which works out to a per-SNP genotyping cost that Chromosomal Address
is a small fraction of a penny. The SNP loci analyzed on A generally useful strategy to identify the defects caus-
commercially available microarrays include all single- ing hereditary diseases is called positional cloning
nucleotide variants known to be associated with genetic (Fig. 11.18). The object is to obtain information about the
diseases, but most of the loci on the chip are common SNPs unknown location of the disease gene by finding poly-
likely to be without phenotypic effect. The widespread oc- morphic loci to which the mutation is genetically linked.
currence of these particular anonymous SNPs makes them Because we know from the human genome sequence the
invaluable for locating the mutations that do cause dis- exact position of each locus, discovering anonymous
eases, as will be explained in the next section. DNA polymorphisms closely linked to the disease gene
allows researchers to focus their search for the mutation
on a small region of a single chromosome. From the
essential concepts candidate genes within this region, the gene responsible
for the disease can be found by looking for mutations that
• In DNA fingerprinting, genotyping of multiple polymorphic appear consistently in patients.
loci such as SSRs provides enough information to identify
individuals from their DNA.
• A DNA microarray contains allele-specific oligonucleotides
(ASOs) for millions of SNP loci. Under the proper
conditions, a probe made of fluorescently-labeled
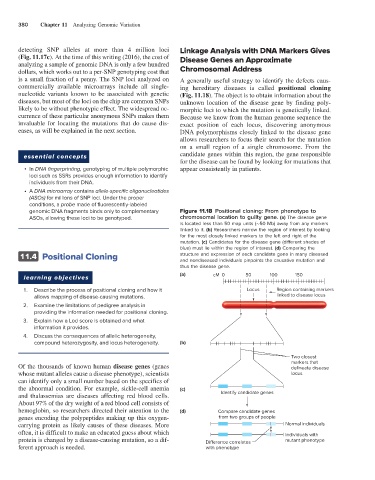
genomic DNA fragments binds only to complementary Figure 11.18 Positional cloning: From phenotype to
ASOs, allowing these loci to be genotyped. chromosomal location to guilty gene. (a) The disease gene
is located less than 50 map units (∼50 Mb) away from any markers
linked to it. (b) Researchers narrow the region of interest by looking
for the most closely linked markers to the left and right of the
mutation. (c) Candidates for the disease gene (different shades of
blue) must lie within the region of interest. (d) Comparing the
11.4 Positional Cloning structure and expression of each candidate gene in many diseased
and nondiseased individuals pinpoints the causative mutation and
thus the disease gene.
(a) cM 0 50 100 150
learning objectives
1. Describe the process of positional cloning and how it Locus Region containing markers
allows mapping of disease-causing mutations. linked to disease locus
2. Examine the limitations of pedigree analysis in
providing the information needed for positional cloning.
3. Explain how a Lod score is obtained and what
information it provides.
4. Discuss the consequences of allelic heterogeneity,
compound heterozygosity, and locus heterogeneity. (b)
Two closest
markers that
Of the thousands of known human disease genes (genes delineate disease
whose mutant alleles cause a disease phenotype), scientists locus
can identify only a small number based on the specifics of
the abnormal condition. For example, sickle-cell anemia (c)
and thalassemias are diseases affecting red blood cells. Identify candidate genes
About 97% of the dry weight of a red blood cell consists of
hemoglobin, so researchers directed their attention to the (d) Compare candidate genes
genes encoding the polypeptides making up this oxygen- from two groups of people
carrying protein as likely causes of these diseases. More Normal individuals
often, it is difficult to make an educated guess about which Individuals with
protein is changed by a disease-causing mutation, so a dif- Di erence correlates mutant phenotype
ferent approach is needed. with phenotype