Page 57 - Genetics_From_Genes_to_Genomes_6th_FULL_Part3
P. 57
10.2 Genome Architecture and Evolution 351
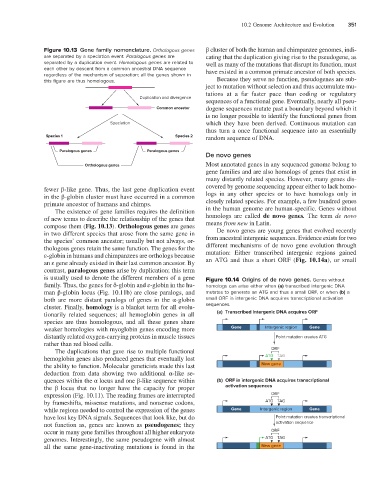
Figure 10.13 Gene family nomenclature. Orthologous genes β cluster of both the human and chimpanzee genomes, indi-
are separated by a speciation event. Paralogous genes are cating that the duplication giving rise to the pseudogene, as
separated by a duplication event. Homologous genes are related to well as many of the mutations that disrupt its function, must
each other by descent from a common ancestral DNA sequence have existed in a common primate ancestor of both species.
regardless of the mechanism of separation; all the genes shown in
this figure are thus homologous. Because they serve no function, pseudogenes are sub-
ject to mutation without selection and thus accumulate mu-
tations at a far faster pace than coding or regulatory
Duplication and divergence
sequences of a functional gene. Eventually, nearly all pseu-
Common ancestor dogene sequences mutate past a boundary beyond which it
is no longer possible to identify the functional genes from
Speciation which they have been derived. Continuous mutation can
thus turn a once functional sequence into an essentially
Species 1 Species 2 random sequence of DNA.
Paralogous genes Paralogous genes
De novo genes
Orthologous genes Most annotated genes in any sequenced genome belong to
gene families and are also homologs of genes that exist in
many distantly related species. However, many genes dis-
fewer β-like gene. Thus, the last gene duplication event covered by genome sequencing appear either to lack homo-
in the β-globin cluster must have occurred in a common logs in any other species or to have homologs only in
primate ancestor of humans and chimps. closely related species. For example, a few hundred genes
The existence of gene families requires the definition in the human genome are human-specific. Genes without
of new terms to describe the relationship of the genes that homologs are called de novo genes. The term de novo
compose them (Fig. 10.13). Orthologous genes are genes means from new in Latin.
in two different species that arose from the same gene in De novo genes are young genes that evolved recently
the species’ common ancestor; usually but not always, or- from ancestral intergenic sequences. Evidence exists for two
thologous genes retain the same function. The genes for the different mechanisms of de novo gene evolution through
ε-globin in humans and chimpanzees are orthologs because mutation: Either transcribed intergenic regions gained
an ε gene already existed in their last common ancestor. By an ATG and thus a short ORF (Fig. 10.14a), or small
contrast, paralogous genes arise by duplication; this term
is usually used to denote the different members of a gene Figure 10.14 Origins of de novo genes. Genes without
family. Thus, the genes for δ-globin and ε-globin in the hu- homologs can arise either when (a) transcribed intergenic DNA
man β-globin locus (Fig. 10.11b) are close paralogs, and mutates to generate an ATG and thus a small ORF, or when (b) a
both are more distant paralogs of genes in the α-globin small ORF in intergenic DNA acquires transcriptional activation
cluster. Finally, homology is a blanket term for all evolu- sequences.
tionarily related sequences; all hemoglobin genes in all (a) Transcribed intergenic DNA acquires ORF
species are thus homologous, and all these genes share
weaker homologies with myoglobin genes encoding more Gene Intergenic region Gene
distantly related oxygen-carrying proteins in muscle tissues Point mutation creates ATG
rather than red blood cells.
The duplications that gave rise to multiple functional ORF
hemoglobin genes also produced genes that eventually lost ATG TAG
the ability to function. Molecular geneticists made this last New gene
deduction from data showing two additional α-like se-
quences within the α locus and one β-like sequence within (b) ORF in intergenic DNA acquires transcriptional
the β locus that no longer have the capacity for proper activation sequences
expression (Fig. 10.11). The reading frames are interrupted ORF
by frameshifts, missense mutations, and nonsense codons, ATGTAG
while regions needed to control the expression of the genes Gene Intergenic region Gene
have lost key DNA signals. Sequences that look like, but do Point mutation creates transcriptional
not function as, genes are known as pseudogenes; they activation sequence
occur in many gene families throughout all higher eukaryote ORF
genomes. Interestingly, the same pseudogene with almost ATGTAG
all the same gene- inactivating mutations is found in the New gene