Page 58 - Genetics_From_Genes_to_Genomes_6th_FULL_Part3
P. 58
352 Chapter 10 Genome Annotation
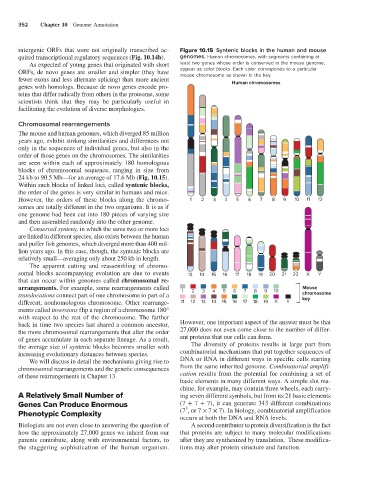
intergenic ORFs that were not originally transcribed ac- Figure 10.15 Syntenic blocks in the human and mouse
quired transcriptional regulatory sequences (Fig. 10.14b). genomes. Human chromosomes, with segments containing at
As expected of young genes that originated with short least two genes whose order is conserved in the mouse genome,
ORFs, de novo genes are smaller and simpler (they have appear as color blocks. Each color corresponds to a particular
mouse chromosome as shown in the key.
fewer exons and less alternate splicing) than more ancient Human chromosomes
genes with homologs. Because de novo genes encode pro-
teins that differ radically from others in the proteome, some
scientists think that they may be particularly useful in
facilitating the evolution of diverse morphologies.
Chromosomal rearrangements
The mouse and human genomes, which diverged 85 million
years ago, exhibit striking similarities and differences not
only in the sequences of individual genes, but also in the
order of those genes on the chromosomes. The similarities
are seen within each of approximately 180 homologous
blocks of chromosomal sequence, ranging in size from
24 kb to 90.5 Mb—for an average of 17.6 Mb (Fig. 10.15).
Within such blocks of linked loci, called syntenic blocks,
the order of the genes is very similar in humans and mice.
However, the orders of these blocks along the chromo- 1 2 3 4 5 6 7 8 9 10 11 12
somes are totally different in the two organisms. It is as if
one genome had been cut into 180 pieces of varying size
and then assembled randomly into the other genome.
Conserved synteny, in which the same two or more loci
are linked in different species, also exists between the human
and puffer fish genomes, which diverged more than 400 mil-
lion years ago. In this case, though, the syntenic blocks are
relatively small—averaging only about 250 kb in length.
The apparent cutting and reassembling of chromo-
somal blocks accompanying evolution are due to events 13 14 15 16 17 18 19 20 21 22 X Y
that can occur within genomes called chromosomal re
arrangements. For example, some rearrangements called 1 2 3 4 5 6 7 8 9 10 Mouse
translocations connect part of one chromosome to part of a chromosome
key
different, nonhomologous chromosome. Other rearrange- 11 12 13 14 15 16 17 18 19 X Y
ments called inversions flip a region of a chromosome 180°
with respect to the rest of the chromosome. The farther
back in time two species last shared a common ancestor, However, one important aspect of the answer must be that
the more chromosomal rearrangements that alter the order 27,000 does not even come close to the number of differ-
of genes accumulate in each separate lineage. As a result, ent proteins that our cells can form.
the average size of syntenic blocks becomes smaller with The diversity of proteins results in large part from
increasing evolutionary distances between species. combinatorial mechanisms that put together sequences of
We will discuss in detail the mechanisms giving rise to DNA or RNA in different ways in specific cells starting
chromosomal rearrangements and the genetic consequences from the same inherited genome. Combinatorial amplifi-
of these rearrangements in Chapter 13. cation results from the potential for combining a set of
basic elements in many different ways. A simple slot ma-
chine, for example, may contain three wheels, each carry-
A Relatively Small Number of ing seven different symbols, but from its 21 basic elements
Genes Can Produce Enormous (7 + 7 + 7), it can generate 343 different combinations
3
Phenotypic Complexity (7 , or 7 × 7 × 7). In biology, combinatorial amplification
occurs at both the DNA and RNA levels.
Biologists are not even close to answering the question of A second contributor to protein diversification is the fact
how the approximately 27,000 genes we inherit from our that proteins are subject to many molecular modifications
parents contribute, along with environmental factors, to after they are synthesized by translation. These modifica-
the staggering sophistication of the human organism. tions may alter protein structure and function.