Page 52 - Genetics_From_Genes_to_Genomes_6th_FULL_Part3
P. 52
346 Chapter 10 Genome Annotation
should also understand that the cDNA library made from red transcribed regions (genes). The idea is very simple: You
blood cell precursors would contain many clones correspond- would determine the sequences of many cDNA clones, and
ing to mRNAs that are highly expressed in this tissue, but then compare these cDNA sequences with that of the ge-
only a few clones that reflect genes that are expressed rarely. nome. Regions of identity between cDNA and genome repre-
sent the exons of genes, and the sequence of a complete
Genomic versus cDNA libraries cDNA (copied from a full-length mRNA) allows you to de-
Figure 10.5 compares genomic and cDNA libraries. The termine the exon/intron structure of the corresponding gene.
clones within genomic libraries represent all regions of Although the basic idea of annotating genomes by com-
DNA equally and show what the intact genome looks like paring cDNA and genomic sequences is straightforward,
in the region of each clone. The clones in cDNA libraries putting it into practice on a large scale is not trivial. Because
reveal which parts of the genome contain the information certain genes are expressed only rarely or only in certain tis-
used in making proteins in specific tissues. The prevalence sue types, genomic scientists need to sequence millions of
of the mRNAs for specific genes also gives some indica- cDNA clones in multiple cDNA libraries made from mRNA
tion, though imperfect, of the relative amounts of the vari- derived from many diverse types of tissue. For this reason, it
ous proteins made in those cells. is likely that some infrequently expressed genes may not yet
As described previously, one of the main purposes of be recognized as genes in genome databases.
making cDNA libraries is to annotate genomes by finding
cDNAs and alternative splicing
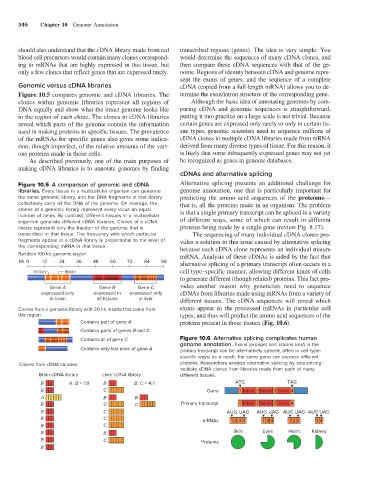
Figure 10.5 A comparison of genomic and cDNA Alternative splicing presents an additional challenge for
libraries. Every tissue in a multicellular organism can generate genome annotation, one that is particularly important for
the same genomic library, and the DNA fragments in that library predicting the amino acid sequences of the proteome—
collectively carry all the DNA of the genome. On average, the that is, all the proteins made in an organism. The problem
clones of a genomic library represent every locus an equal is that a single primary transcript can be spliced in a variety
number of times. By contrast, different tissues in a multicellular
organism generate different cDNA libraries. Clones of a cDNA of different ways, some of which can result in different
library represent only the fraction of the genome that is proteins being made by a single gene (review Fig. 8.17).
transcribed in that tissue. The frequency with which particular The sequencing of many individual cDNA clones pro-
fragments appear in a cDNA library is proportional to the level of vides a solution to this issue caused by alternative splicing
the corresponding mRNA in that tissue. because each cDNA clone represents an individual mature
Random 100 kb genomic region mRNA. Analysis of these cDNAs is aided by the fact that
kb 0 12 24 36 48 60 72 84 96 alternative splicing of a primary transcript often occurs in a
Intron Exon cell type–specific manner, allowing different kinds of cells
to generate different (though related) proteins. This fact pro-
Gene A Gene B Gene C vides another reason why geneticists need to sequence
expressed only expressed in expressed only cDNAs from libraries made using mRNAs from a variety of
in brain all tissues in liver different tissues. The cDNA sequences will reveal which
Clones from a genomic library with 20 kb inserts that come from exons appear in the processed mRNAs in particular cell
this region types, and thus will predict the amino acid sequences of the
Contains part of gene A proteins present in those tissues (Fig. 10.6).
Contains parts of genes B and C
Contains all of gene C Figure 10.6 Alternative splicing complicates human
genome annotation. Exons (orange) and introns (red) in the
Contains only last exon of gene A primary transcript can be alternatively spliced, often in cell type-
specific ways; as a result, the same gene can express different
Clones from cDNA libraries proteins. Researchers analyze alternative splicing by sequencing
multiple cDNA clones from libraries made from each of many
Brain cDNA library Liver cDNA library different tissues.
B A : B = 1:9 B B :C = 4:7 ATG TAG
B C Gene 1 Intron 2 Intron 3 Intron 4
A B B
B C C Primary transcript 1 Intron 2 Intron 3 Intron 4
B C AUG UAG AUG UAG AUG UAG AUG UAG
B C mRNAs 12 34 13 4 12 4 1 4
B C
B B Skin Eyes Heart Kidney
B C Proteins
B