Page 51 - Genetics_From_Genes_to_Genomes_6th_FULL_Part3
P. 51
10.1 Finding the Genes in Genomes 345
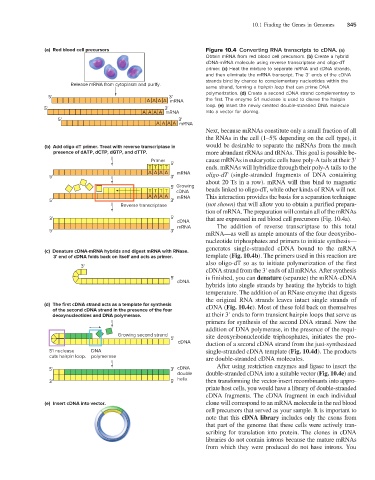
(a) Red blood cell precursors Figure 10.4 Converting RNA transcripts to cDNA. (a)
Obtain mRNA from red blood cell precursors. (b) Create a hybrid
cDNA-mRNA molecule using reverse transcriptase and oligo-dT
primer. (c) Heat the mixture to separate mRNA and cDNA strands,
and then eliminate the mRNA transcript. The 3′ ends of the cDNA
strands bind by chance to complementary nucleotides within the
Release mRNA from cytoplasm and purify.
same strand, forming a hairpin loop that can prime DNA
polymerization. (d) Create a second cDNA strand complementary to
5' 3'
A AAA mRNA the first. The enzyme S1 nuclease is used to cleave the hairpin
5' 3' loop. (e) Insert the newly created double-stranded DNA molecule
AA AA mRNA into a vector for cloning.
5' 3'
AA AA mRNA
Next, because mRNAs constitute only a small fraction of all
the RNAs in the cell (1–5% depending on the cell type), it
(b) Add oligo-dT primer. Treat with reverse transcriptase in would be desirable to separate the mRNAs from the much
presence of dATP, dCTP, dGTP, and dTTP. more abundant rRNAs and tRNAs. This goal is possible be-
Primer 5' cause mRNAs in eukaryotic cells have poly-A tails at their 3′
TTT T ends. mRNAs will hybridize through their poly-A tails to the
AA AA mRNA oligo-dT (single-stranded fragments of DNA containing
5' 3'
about 20 Ts in a row). mRNA will thus bind to magnetic
5' Growing beads linked to oligo-dT, while other kinds of RNA will not.
TTT T cDNA
AA AA mRNA This interaction provides the basis for a separation technique
5' 3'
Reverse transcriptase (not shown) that will allow you to obtain a purified prepara-
tion of mRNA. The preparation will contain all of the mRNAs
3' 5' that are expressed in red blood cell precursors (Fig. 10.4a).
cDNA
mRNA The addition of reverse transcriptase to this total
5' 3' mRNA—as well as ample amounts of the four deoxyribo-
nucleotide triphosphates and primers to initiate synthesis—
generates single-stranded cDNA bound to the mRNA
(c) Denature cDNA-mRNA hybrids and digest mRNA with RNase.
3' end of cDNA folds back on itself and acts as primer. template (Fig. 10.4b). The primers used in this reaction are
also oligo-dT so as to initiate polymerization of the first
3'
cDNA strand from the 3′ ends of all mRNAs. After synthesis
5' is finished, you can denature (separate) the mRNA-cDNA
cDNA
hybrids into single strands by heating the hybrids to high
temperature. The addition of an RNase enzyme that digests
the original RNA strands leaves intact single strands of
(d) The first cDNA strand acts as a template for synthesis cDNA (Fig. 10.4c). Most of these fold back on themselves
of the second cDNA strand in the presence of the four
deoxynucleotides and DNA polymerase. at their 3′ ends to form transient hairpin loops that serve as
primers for synthesis of the second DNA strand. Now the
addition of DNA polymerase, in the presence of the requi-
Growing second strand site deoxyribonucleotide triphosphates, initiates the pro-
5'
cDNA duction of a second cDNA strand from the just-synthesized
S1 nuclease DNA single-stranded cDNA template (Fig. 10.4d). The products
cuts hairpin loop. polymerase are double-stranded cDNA molecules.
5' 3' cDNA After using restriction enzymes and ligase to insert the
double double-stranded cDNA into a suitable vector (Fig. 10.4e) and
3' 5' helix then transforming the vector-insert recombinants into appro-
priate host cells, you would have a library of double-stranded
cDNA fragments. The cDNA fragment in each individual
(e) Insert cDNA into vector. clone will correspond to an mRNA molecule in the red blood
cell precursors that served as your sample. It is important to
note that this cDNA library includes only the exons from
that part of the genome that these cells were actively tran-
scribing for translation into protein. The clones in cDNA
libraries do not contain introns because the mature mRNAs
from which they were produced do not have introns. You