Page 42 - Genetics_From_Genes_to_Genomes_6th_FULL_Part3
P. 42
336 Chapter 9 Digital Analysis of DNA
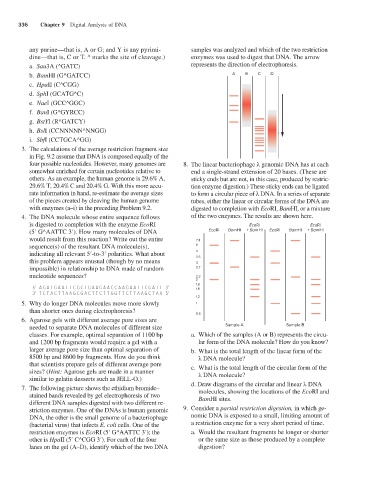
any purine—that is, A or G; and Y is any pyrimi- samples was analyzed and which of the two restriction
dine—that is, C or T. ^ marks the site of cleavage.) enzymes was used to digest that DNA. The arrow
a. Sau3A (^GATC) represents the direction of electrophoresis.
b. BamHI (G^GATCC) A B C D
c. HpaII (C^CGG)
d. SphI (GCATG^C)
e. NaeI (GCC^GGC)
f. BanI (G^GYRCC)
g. BstYI (R^GATCY)
h. BslI (CCNNNNN^NNGG)
i. SbfI (CCTGCA^GG)
3. The calculations of the average restriction fragment size
in Fig. 9.2 assume that DNA is composed equally of the
four possible nucleotides. However, many genomes are 8. The linear bacteriophage λ genomic DNA has at each
somewhat enriched for certain nucleotides relative to end a single-strand extension of 20 bases. (These are
others. As an example, the human genome is 29.6% A, sticky ends but are not, in this case, produced by restric-
29.6% T, 20.4% C and 20.4% G. With this more accu- tion enzyme digestion.) These sticky ends can be ligated
rate information in hand, re-estimate the average sizes to form a circular piece of λ DNA. In a series of separate
of the pieces created by cleaving the human genome tubes, either the linear or circular forms of the DNA are
with enzymes (a–i) in the preceding Problem 9.2. digested to completion with EcoRI, BamHI, or a mixture
4. The DNA molecule whose entire sequence follows of the two enzymes. The results are shown here.
is digested to completion with the enzyme EcoRI EcoRI EcoRI
(5′ G^AATTC 3′). How many molecules of DNA EcoRI BamHI + BamHI EcoRI BamHI + BamHI
would result from this reaction? Write out the entire 7.3
sequence(s) of the resultant DNA molecule(s), 5
indicating all relevant 5′-to-3′ polarities. What about 4 3.5
this problem appears unusual (though by no means 3
impossible) in relationship to DNA made of random 2.7
nucleotide sequences? 2.2
2
1.8
5' A G A TG AA TT CG CT GA AG AA CC AA GA AT TC GA TT 3' 1.5
3' T C T AC TT AA GC GA CT TC TT GG TT CT TA AG CT AA 5'
1.2
5. Why do longer DNA molecules move more slowly 1
than shorter ones during electrophoresis?
0.5
6. Agarose gels with different average pore sizes are
needed to separate DNA molecules of different size Sample A Sample B
classes. For example, optimal separation of 1100 bp a. Which of the samples (A or B) represents the circu-
and 1200 bp fragments would require a gel with a lar form of the DNA molecule? How do you know?
larger average pore size than optimal separation of b. What is the total length of the linear form of the
8500 bp and 8600 bp fragments. How do you think λ DNA molecule?
that scientists prepare gels of different average pore c. What is the total length of the circular form of the
sizes? (Hint: Agarose gels are made in a manner λ DNA molecule?
similar to gelatin desserts such as JELL-O.)
7. The following picture shows the ethidium bromide– d. Draw diagrams of the circular and linear λ DNA
molecules, showing the locations of the EcoRI and
stained bands revealed by gel electrophoresis of two BamHI sites.
different DNA samples digested with two different re-
striction enzymes. One of the DNAs is human genomic 9. Consider a partial restriction digestion, in which ge-
DNA, the other is the small genome of a bacteriophage nomic DNA is exposed to a small, limiting amount of
(bacterial virus) that infects E. coli cells. One of the a restriction enzyme for a very short period of time.
restriction enzymes is EcoRI (5′ G^AATTC 3′); the a. Would the resultant fragments be longer or shorter
other is HpaII (5′ C^CGG 3′). For each of the four or the same size as those produced by a complete
lanes on the gel (A–D), identify which of the two DNA digestion?