Page 63 - Genetics_From_Genes_to_Genomes_6th_FULL_Part2
P. 63
222 Chapter 7 Anatomy and Function of a Gene: Dissection Through Mutation
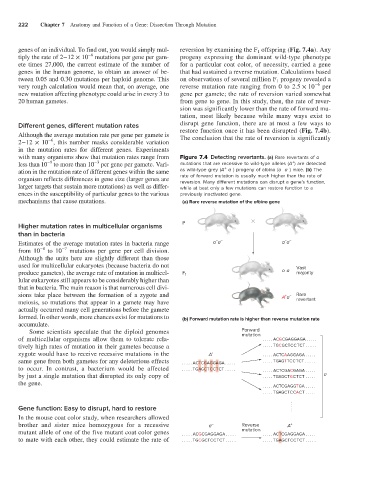
genes of an individual. To find out, you would simply mul- reversion by examining the F 1 offspring (Fig. 7.4a). Any
−6
tiply the rate of 2−12 × 10 mutations per gene per gam- progeny expressing the dominant wild-type phenotype
ete times 27,000, the current estimate of the number of for a particular coat color, of necessity, carried a gene
genes in the human genome, to obtain an answer of be- that had sustained a reverse mutation. Calculations based
tween 0.05 and 0.30 mutations per haploid genome. This on observations of several million F 1 progeny revealed a
−6
very rough calculation would mean that, on average, one reverse mutation rate ranging from 0 to 2.5 × 10 per
new mutation affecting phenotype could arise in every 3 to gene per gamete; the rate of reversion varied somewhat
20 human gametes. from gene to gene. In this study, then, the rate of rever-
sion was significantly lower than the rate of forward mu-
tation, most likely because while many ways exist to
Different genes, different mutation rates disrupt gene function, there are at most a few ways to
Although the average mutation rate per gene per gamete is restore function once it has been disrupted (Fig. 7.4b).
The conclusion that the rate of reversion is significantly
−6
2−12 × 10 , this number masks considerable variation
in the mutation rates for different genes. Experiments
with many organisms show that mutation rates range from Figure 7.4 Detecting revertants. (a) Rare revertants of a
–
−9
−3
+
less than 10 to more than 10 per gene per gamete. Vari- mutations that are recessive to wild-type alleles (A ) are detected
–
+
–
–
ation in the mutation rate of different genes within the same as wild-type grey (A a ) progeny of albino (a a ) mice. (b) The
organism reflects differences in gene size (larger genes are rate of forward mutation is usually much higher than the rate of
reversion. Many different mutations can disrupt a gene’s function,
larger targets that sustain more mutations) as well as differ- while at best only a few mutations can restore function to a
ences in the susceptibility of particular genes to the various previously inactivated gene.
mechanisms that cause mutations. (a) Rare reverse mutation of the albino gene
Higher mutation rates in multicellular organisms P
than in bacteria
− −
− −
Estimates of the average mutation rates in bacteria range a a a a
−7
−8
from 10 to 10 mutations per gene per cell division.
Although the units here are slightly different than those
used for multicellular eukaryotes (because bacteria do not Vast
− −
produce gametes), the average rate of mutation in multicel- F 1 a a majority
lular eukaryotes still appears to be considerably higher than
that in bacteria. The main reason is that numerous cell divi-
sions take place between the formation of a zygote and A a Rare
+ −
meiosis, so mutations that appear in a gamete may have revertant
actually occurred many cell generations before the gamete
formed. In other words, more chances exist for mutations to (b) Forward mutation rate is higher than reverse mutation rate
accumulate.
Some scientists speculate that the diploid genomes Forward
of multicellular organisms allow them to tolerate rela- mutation . . . . . ACGCGAGGAGA . . . . .
tively high rates of mutation in their gametes because a . . . . . TGCGCTCCTCT . . . . .
zygote would have to receive recessive mutations in the A + . . . . . ACTCAAGGAGA . . . . .
same gene from both gametes for any deleterious effects . . . . . ACTCGAGGAGA . . . . . . . . . . TGAGTTCCTCT . . . . .
to occur. In contrast, a bacterium would be affected . . . . . TGAGCTCCTCT . . . . . . . . . . ACTCGACGAGA . . . . .
by just a single mutation that disrupted its only copy of . . . . . TGAGCTGCTCT . . . . . a −
the gene.
. . . . . ACTCGAGGTGA . . . . .
. . . . . TGAGCTCCACT . . . . .
Gene function: Easy to disrupt, hard to restore . . . . .
In the mouse coat color study, when researchers allowed
brother and sister mice homozygous for a recessive a − Reverse A +
mutant allele of one of the five mutant coat color genes . . . . . ACGCGAGGAGA . . . . . mutation . . . . . ACTCGAGGAGA . . . . .
to mate with each other, they could estimate the rate of . . . . . TGCGCTCCTCT . . . . . . . . . . TGAGCTCCTCT . . . . .