Page 16 - Genetics_From_Genes_to_Genomes_6th_FULL_Part1
P. 16
What’s Next
GENETICS AND SOCIETY
© Image Source/Getty Images RF
Should We Alter Human Germ-Line Genomes?
In April 2015, Chinese scientists reported the use of CRISPR/
but it can cause unwanted off-target effects that alter sequences
Cas9 to correct ß-globin gene mutations—the cause of the dis-
ease ß-thalassemia—in human embryos. Although these em-
elsewhere in the genome. The consequences of these off-tar-
bryos were never placed in a womb, this publication opened a
get mutations when transmitted over many generations are un-
predictable; this is why the international summit concluded the
firestorm of controversy because some descendants of embry-
onic cells eventually will become sperm or eggs that could be
method is premature.
But even if the technologies can be perfected, should we
passed down to future generations. In other words, these stud-
ies demonstrated forcefully that gene editing technology is be-
ever employ them to alter human germ lines in eggs, sperm, or
embryos? Some people believe the entire idea is unethical be-
coming powerful enough that humans will soon be able to
cause decisions made now will impact our descendants without
change their own evolutionary destiny.
their consent. Because it is conceivable that genome modifica-
In response to this report, the governments of the United
tions can be made eventually that will enhance traits like intelli-
States, the United Kingdom, and China organized an interna-
tional summit on human gene editing, held in Washington, D.C.
gence, some people argue that germ-line editing technologies
will inevitably lead to a further stratification of society: Likely,
in December of 2015 and attended by more than 500 scientists,
only wealthy individuals would be able to afford to have
ethicists, and legal experts from 20 countries. The strong con-
“designer children” with these enhanced characteristics. But on
sensus of the summit was that genome editing of human em-
bryos intended for pregnancy is premature because its safety
the other side of the issue, some scientists argue that if gene
editing can be shown to be safe, without off-target effects, it
cannot be ensured, but the participants were divided as to
would be unethical not to use this technology, at least to eradi-
whether the goal of altering human germ lines is ethical or desir-
able. As of this writing in 2016, the British and Chinese govern-
cate disease if not to improve human traits.
ments are likely to continue to fund research involving genome
Genome editing methods are advancing so rapidly that
these issues will soon go beyond interesting theoretical debates
editing of human embryos not destined for pregnancy, but in the
to the point where they have real impact on people’s lives and
United States only private agencies fund such investigations.
Gene editing of somatic cells to cure the symptoms of
those of future generations. If mankind will intentionally alter its
disease is relatively noncontroversial, but altering germ-line
own evolution, we had better be sure that the vast potential impli-
genomes raises many issues. Some of these issues are
cations of these decisions have been thoroughly considered.
• Therapeutic genes can be delivered in recombinant viral
• DNA introduced in adenoviral vectors remains extrachro-
mosomal, necessitating periodic repeats of the therapy.
vectors to somatic cells of patients either in vivo or ex vivo.
• Scientists are gearing up to use genome editing methods
• Retroviral vectors insert therapy genes into human
such as CRISPR/Cas9 to repair mutant genes in human
chromosomes, but this method can result in gene
somatic cells.
mutation and cancer.
essential concepts technical. For example, CRISPR/Cas9 technology is powerful, 637 Guided Tour xv
210 Chapter 6 DNA Structure, Replication, and Recombination
DNA sequences. Site-specific recombination is crossing- between two target sites in a single chromosome is removed
over that occurs only between to create two independent DNA molecules (Fig. 6.31b). If
WHAT’S NEXT two specific DNA target
What’s Next
sites that are usually less than 200 base pairs long. Site- a bacteriophage genome was previously integrated into the
specific recombination is much simpler at the molecular
host chromosome, excision is crucial to allow the bacterio-
Manipulation of the genome is the basis for many of the ex- complex multicellular organism. Transgenic technology is Each chapter closes with a What’s
level than is the homologous recombination discussed in
perimental strategies we will describe in Chapter 19, where key to cloning the genes identified in mutant screens that Next section that serves as a bridge
phage genome to extricate itself and then to become incor-
we discuss how genetic analysis has been a crucial tool in systems of porated in the virus particle.
the previous section. In particular, in most
are crucial for regulating development, and also to manipu-
unraveling the biochemical pathways of development—the lating these genes in order to understand their precise func- between the topics in the chapter just
site-specific recombination, a single protein logically
A third potential outcome of site-specific recombina-
process by which a single-celled zygote becomes a tions in the organism. completed to those in the upcoming
called a recombinase is sufficient to catalyze all the
chapter or chapters. This spirals the
DNA: © Design Pics/Bilderbuch RF tion systems is the inversion of a segment of DNA that is
breakage and joining steps of the process. If you are curi- located between the two target sites (Fig. 6.31c). As you
learning and builds connections for
ous, Fig. 6.30 depicts the mode of action of one class of can imagine, such inversion could constitute a molecular
students.
such recombinases. switch between two configurations of the same chromo-
The organisms that take advantage of site-specific some. The in-between segment is oriented in one direction
recombination include certain kinds of bacteriophages
har00909_ch18_618-645.indd 637 in one state and in the other direction in the other state.
6/13/17 7:56 PM
that use this process for the integration (incorporation) A final mode of site-specific recombination can occur
of their small, circular genome into the chromosome of if the target site is found at the same position on each of two
New! Exciting Revised Content
the host bacterium (Fig. 6.31a). In this way, the bacteri- homologous chromosomes. Action of the recombinase on
ophage DNA “hitchhikes” along with the bacterial chro- these target sites will result in the reshuffling of regions on
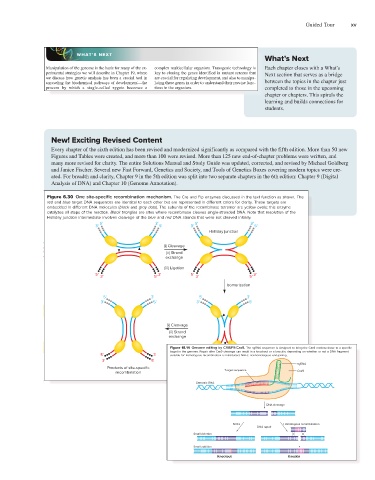
Every chapter of the sixth edition has been revised and modernized significantly as compared with the fifth edition. More than 50 new
mosome: When the host DNA replicates, so does the nonsister chromatids, an outcome that leads to recombinant
Figures and Tables were created, and more than 100 were revised. More than 125 new end-of-chapter problems were written, and
integrated bacteriophage genome. chromosomes (Fig. 6.31d). To our knowledge, this situa-
many more revised for clarity. The entire Solutions Manual and Study Guide was updated, corrected, and revised by Michael Goldberg
Site-specific recombination is also important for the
tion is not normally encountered in organisms that natu-
and Janice Fischer. Several new Fast Forward, Genetics and Society, and Tools of Genetics Boxes covering modern topics were cre-
reverse process of excision, in which the DNA integrated rally use site-specific recombination. However, geneticists
ated. For breadth and clarity, Chapter 9 in the 5th edition was split into two separate chapters in the 6th edition: Chapter 9 (Digital
Analysis of DNA) and Chapter 10 (Genome Annotation).
Figure 6.30 One site-specific recombination mechanism. The Cre and Flp enzymes discussed in the text function as shown. The
red and blue target DNA sequences are identical to each other but are represented in different colors for clarity. These targets are
Chapter 18 Manipulating the Genomes of Eukaryotes
632
embedded in different DNA molecules (black and gray dots). The subunits of the recombinase tetramer are yellow ovals; this enzyme
catalyzes all steps of the reaction. Black triangles are sites where recombinase cleaves single-stranded DNA. Note that resolution of the
mutant mice more efficiently. Of wider importance,
Holliday junction intermediate involves cleavage of the blue and red DNA strands that were not cleaved initially. single-stranded RNA called sgRNA (single guide RNA).
3'
5' 3' researchers can apply the same tools in animals other than At the 5′ end of the sgRNA is a 20 bp sequence that is
5'
3' 5' mice, or even in cultured cells, opening up many possibilities complementary in sequence to a target site of interest in
5'
3'
for the study of gene function and to establish new models the genome to be altered. The 3′ end of the sgRNA binds
Holliday junction
for human diseases. specifically to the Cas9 protein. (As an aside, the 5′ and
In all of these technologies, either a protein or an RNA 3′ regions of the sgRNA correspond respectively to the
molecule serves as a guide that brings a DNA-cleaving en-
(i) Cleavage crRNA and tracrRNA in the Tools of Genetics Box.) The
zyme to a specific genomic location. DNA repair of the second component is a Cas9 polypeptide that has been
break can then result in a point mutation (a base pair change,
(ii) Strand altered so that it includes a short stretch of amino acids
exchange that constitute a nuclear localization signal, allowing the
or insertion or deletion of one or a few pairs) or a knockin of
specific DNA sequences. We describe here the newest and protein to be imported into the nucleus where it can act
most efficient genome editing system, called CRISPR/Cas9.
(iii) Ligation on DNA.
CRISPR is an acronym for clustered regularly inter- In the nucleus, Cas9/sgRNA complexes seek out and
3'
5' 3' 5' 3' spaced short palindromic repeats. Many bacterial genomes bind to their designated genomic DNA target. The Cas9
5'
3'
5'
contain a CRISPR region, which functions as an antiviral enzyme within the complexes then makes a double-strand
immune system. CRISPR immunity also depends on endo- break in the target DNA (Fig. 18.14). Repair of the break
Isomerization
nucleases called Cas proteins (CRISPR-associated proteins) by nonhomologous end-joining (NHEJ; review Fig. 7.18)
encoded by the bacterial genome; these enzymes can make often results in a small insertion or a deletion of a few base
5'
5' 3' double-stranded breaks in DNA. The Tools of Genetics Box pairs at the break. Such a mutation can knockout the func-
3'
3' 5' entitled How Bacteria Vaccinate Themselves Against tion of a gene, for example if it corresponds to a frameshift
3'
5' Viral
Infections with CRISPR/Cas9 describes in detail how bacte- mutation in an open reading frame.
ria use this mechanism to ward off infection by bacterio- Alternatively, if DNA molecules corresponding to the
phages. The attention of the scientific community became DNA flanking the break are introduced into cells at the
focused on CRISPR/Cas9 when researchers realized they same time as the Cas9/sgRNA, double-strand break repair
could adapt this system for use in any organism.
(i) Cleavage by homologous recombination can incorporate that DNA
The genetically engineered CRISPR/Cas9 system has into the genome at the break site, generating a knockin
(ii) Strand (Fig. 18.14). Double-strand breaks are recombinogenic
two components. The first is an investigator-designed,
exchange
Figure 18.14 Genome editing by CRISPR/Cas9. The sgRNA sequence is designed to bring the Cas9 endonuclease to a specific
(iii) Ligation
target in the genome. Repair after Cas9 cleavage can result in a knockout or a knockin, depending on whether or not a DNA fragment
5' 3' suitable for homologous recombination is introduced. NHEJ: 3' nonhomologous end-joining.
5'
3' 5' 3' 5'
sgRNA
Products of site-specific
recombination Target sequence Cas9
Genomic DNA
DNA cleavage
har00909_ch06_181-218.indd 210 5/12/17 9:21 PM
NHEJ Homologous recombination
DNA repair
Small deletion
Small addition
Knockout Knockin
har00909_ch18_618-645.indd 632 6/30/17 10:53 AM