Page 129 - Genetics_From_Genes_to_Genomes_6th_FULL_Part1
P. 129
4.7 Sex-Linked and Sexually Dimorphic Traits in Humans 121
FAST FORWARD
Visualizing X Chromosome Inactivation in Transgenic Mice
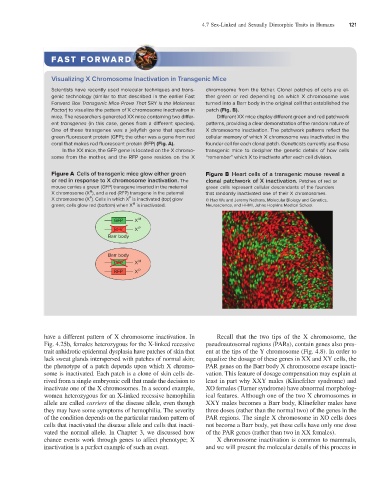
Scientists have recently used molecular techniques and trans- chromosome from the father. Clonal patches of cells are ei-
genic technology (similar to that described in the earlier Fast ther green or red depending on which X chromosome was
Forward Box Transgenic Mice Prove That SRY Is the Maleness turned into a Barr body in the original cell that established the
Factor) to visualize the pattern of X chromosome inactivation in patch (Fig. B).
mice. The researchers generated XX mice containing two differ- Different XX mice display different green and red patchwork
ent transgenes (in this case, genes from a different species). patterns, providing a clear demonstration of the random nature of
One of these transgenes was a jellyfish gene that specifies X chromosome inactivation. The patchwork patterns reflect the
green fluorescent protein (GFP); the other was a gene from red cellular memory of which X chromosome was inactivated in the
coral that makes red fluorescent protein (RFP) (Fig. A). founder cell for each clonal patch. Geneticists currently use these
In the XX mice, the GFP gene is located on the X chromo- transgenic mice to decipher the genetic details of how cells
some from the mother, and the RFP gene resides on the X “remember” which X to inactivate after each cell division.
Figure A Cells of transgenic mice glow either green Figure B Heart cells of a transgenic mouse reveal a
or red in response to X chromosome inactivation. The clonal patchwork of X inactivation. Patches of red or
mouse carries a green (GFP) transgene inserted in the maternal green cells represent cellular descendants of the founders
M
X chromosome (X ), and a red (RFP) transgene in the paternal that randomly inactivated one of their X chromosomes.
P
P
X chromosome (X ). Cells in which X is inactivated (top) glow © Hao Wu and Jeremy Nathans, Molecular Biology and Genetics,
M
green; cells glow red (bottom) when X is inactivated. Neuroscience, and HHMI, Johns Hopkins Medical School.
GFP X M
RFP X P
Barr body
Barr body
GFP X M
RFP X P
have a different pattern of X chromosome inactivation. In Recall that the two tips of the X chromosome, the
Fig. 4.25b, females heterozygous for the X-linked recessive pseudoautosomal regions (PARs), contain genes also pres-
trait anhidrotic epidermal dysplasia have patches of skin that ent at the tips of the Y chromosome (Fig. 4.8). In order to
lack sweat glands interspersed with patches of normal skin; equalize the dosage of these genes in XX and XY cells, the
the phenotype of a patch depends upon which X chromo- PAR genes on the Barr body X chromosome escape inacti-
some is inactivated. Each patch is a clone of skin cells de- vation. This feature of dosage compensation may explain at
rived from a single embryonic cell that made the decision to least in part why XXY males (Klinefelter syndrome) and
inactivate one of the X chromosomes. In a second example, XO females (Turner syndrome) have abnormal morpholog-
women heterozygous for an X-linked recessive hemophilia ical features. Although one of the two X chromosomes in
allele are called carriers of the disease allele, even though XXY males becomes a Barr body, Klinefelter males have
they may have some symptoms of hemophilia. The severity three doses (rather than the normal two) of the genes in the
of the condition depends on the particular random pattern of PAR regions. The single X chromosome in XO cells does
cells that inactivated the disease allele and cells that inacti- not become a Barr body, yet these cells have only one dose
vated the normal allele. In Chapter 3, we discussed how of the PAR genes (rather than two in XX females).
chance events work through genes to affect phenotype; X X chromosome inactivation is common to mammals,
inactivation is a perfect example of such an event. and we will present the molecular details of this process in