Page 24 - Genetics_From_Genes_to_Genomes_6th_FULL_Part3
P. 24
318 Chapter 9 Digital Analysis of DNA
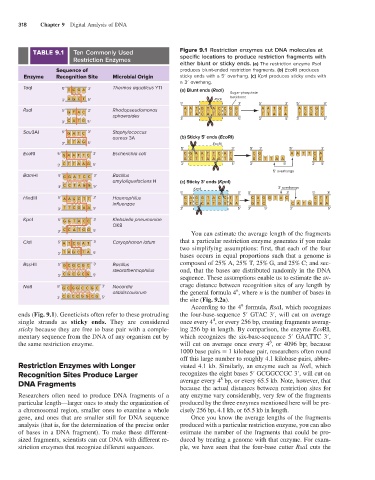
TABLE 9.1 Ten Commonly Used Figure 9.1 Restriction enzymes cut DNA molecules at
Restriction Enzymes specific locations to produce restriction fragments with
either blunt or sticky ends. (a) The restriction enzyme RsaI
Sequence of produces blunt-ended restriction fragments. (b) EcoRI produces
Enzyme Recognition Site Microbial Origin sticky ends with a 5′ overhang. (c) KpnI produces sticky ends with
a 3′ overhang.
TaqI 5' T C G A 3' Thermus aquaticus YTI (a) Blunt ends (RsaI)
Sugar-phosphate
backbone
3' A G C T 5' RsaI
5' 3' 5' 3' 5' 3'
RsaI 5' G TA C 3' Rhodopseudomonas AA T G TA C C G G A A T G T A C C G G
sphaeroides 3' TT A C A T G G C C 5' 3' T T A C A 5' 3' T G G C C 5'
3' C A T G 5'
Sau3AI 5' G A T C 3' Staphylococcus
aureus 3A (b) Sticky 5' ends (EcoRI)
3' C T A G 5' EcoRI
5' 3' 5' 3' 5' 3'
EcoRl 5' G A A T T C 3' Escherichia coli C G A A T T C A T C G A A T T C A
GC T T A A G T A GC T T A A G T
3' C T T A A G 5' 3' 5' 3' 5' 3' 5'
5' overhangs
BamHI 5' G G A T C C 3' Bacillus
amyloliquefaciens H (c) Sticky 3' ends (KpnI)
3' C C T A G G 5' KpnI 3' overhangs
5' 3' 5' 3' 5' 3'
HindIII 5' A A G C T T 3' Haemophilus C A G G TA C C T T C A G G TA C C T T
influenzae G T C C A T G G A A G T C C A T G G A A
3' T T C GA A 5' 3' 5' 3' 5' 3' 5'
KpnI 5' G G TA C C 3' Klebsiella pneumoniae
OK8
3' C C A T G G 5' You can estimate the average length of the fragments
ClaI 5' A T C G A T 3' Caryophanon latum that a particular restriction enzyme generates if you make
two simplifying assumptions: first, that each of the four
3' T A G C T A 5' bases occurs in equal proportions such that a genome is
BssHII 5' G C G G CC 3' Bacillus composed of 25% A, 25% T, 25% G, and 25% C; and sec-
stearothermophilus ond, that the bases are distributed randomly in the DNA
3' C G C G C G 5' sequence. These assumptions enable us to estimate the av-
NotI 5' G C G G C C G C 3' Nocardia erage distance between recognition sites of any length by
n
otitidiscaviarum the general formula 4 , where n is the number of bases in
3' C G C C G G C G 5' the site (Fig. 9.2a).
According to the 4 formula, RsaI, which recognizes
n
ends (Fig. 9.1). Geneticists often refer to these protruding the four-base-sequence 5′ GTAC 3′, will cut on average
4
single strands as sticky ends. They are considered once every 4 , or every 256 bp, creating fragments averag-
sticky because they are free to base pair with a comple- ing 256 bp in length. By comparison, the enzyme EcoRI,
mentary sequence from the DNA of any organism cut by which recognizes the six-base-sequence 5′ GAATTC 3′,
6
the same restriction enzyme. will cut on average once every 4 , or 4096 bp; because
1000 base pairs = 1 kilobase pair, researchers often round
off this large number to roughly 4.1 kilobase pairs, abbre-
Restriction Enzymes with Longer viated 4.1 kb. Similarly, an enzyme such as NotI, which
Recognition Sites Produce Larger recognizes the eight bases 5′ GCGGCCGC 3′, will cut on
8
DNA Fragments average every 4 bp, or every 65.5 kb. Note, however, that
because the actual distances between restriction sites for
Researchers often need to produce DNA fragments of a any enzyme vary considerably, very few of the fragments
particular length—larger ones to study the organization of produced by the three enzymes mentioned here will be pre-
a chromosomal region, smaller ones to examine a whole cisely 256 bp, 4.1 kb, or 65.5 kb in length.
gene, and ones that are smaller still for DNA sequence Once you know the average lengths of the fragments
analysis (that is, for the determination of the precise order produced with a particular restriction enzyme, you can also
of bases in a DNA fragment). To make these different- estimate the number of the fragments that could be pro-
sized fragments, scientists can cut DNA with different re- duced by treating a genome with that enzyme. For exam-
striction enzymes that recognize different sequences. ple, we have seen that the four-base cutter RsaI cuts the