Page 40 - Genetics_From_Genes_to_Genomes_6th_FULL_Part2
P. 40
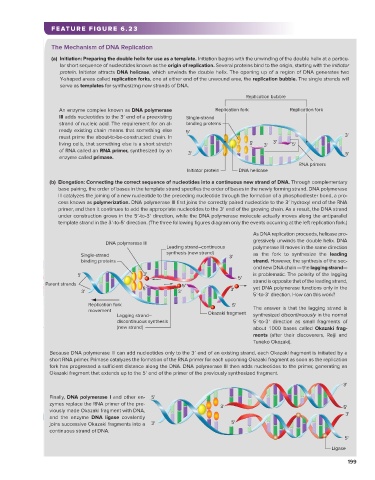
FEATURE FIGURE 6.23
The Mechanism of DNA Replication
(a) Initiation: Preparing the double helix for use as a template. Initiation begins with the unwinding of the double helix at a particu-
lar short sequence of nucleotides known as the origin of replication. Several proteins bind to the origin, starting with the initiator
protein. Initiator attracts DNA helicase, which unwinds the double helix. The opening up of a region of DNA generates two
Y-shaped areas called replication forks, one at either end of the unwound area, the replication bubble. The single strands will
serve as templates for synthesizing new strands of DNA.
Replication bubble
An enzyme complex known as DNA polymerase Replication fork Replication fork
III adds nucleotides to the 3′ end of a preexisting Single-strand
strand of nucleic acid. The requirement for an al- binding proteins
ready existing chain means that something else 5'
must prime the about-to-be-constructed chain. In 3'
living cells, that something else is a short stretch 5' 3' 3' 5'
of RNA called an RNA primer, synthesized by an 3' 5'
enzyme called primase.
RNA primers
Initiator protein DNA helicase
(b) Elongation: Connecting the correct sequence of nucleotides into a continuous new strand of DNA. Through complementary
base pairing, the order of bases in the template strand specifies the order of bases in the newly forming strand. DNA polymerase
III catalyzes the joining of a new nucleotide to the preceding nucleotide through the formation of a phosphodiester bond, a pro-
cess known as polymerization. DNA polymerase III first joins the correctly paired nucleotide to the 3′ hydroxyl end of the RNA
primer, and then it continues to add the appropriate nucleotides to the 3′ end of the growing chain. As a result, the DNA strand
under construction grows in the 5′-to-3′ direction, while the DNA polymerase molecule actually moves along the antiparallel
template strand in the 3′-to-5′ direction. (The three following figures diagram only the events occurring at the left replication fork.)
As DNA replication proceeds, helicase pro-
gressively unwinds the double helix. DNA
DNA polymerase III
Leading strand–continuous polymerase III moves in the same direction
Single-strand synthesis (new strand) 3' as the fork to synthesize the leading
binding proteins strand. However, the synthesis of the sec-
ond new DNA chain — the lagging strand—
5' 3' is problematic. The polarity of the lagging
5' strand is opposite that of the leading strand,
Parent strands 5'
5' 3' yet DNA polymerase functions only in the
3' 3'
5′-to-3′ direction. How can this work?
Replication fork 5' The answer is that the lagging strand is
movement Okazaki fragment
Lagging strand– synthesized discontinuously in the normal
discontinuous synthesis 5′-to-3′ direction as small fragments of
(new strand) about 1000 bases called Okazaki frag-
ments (after their discoverers, Reiji and
Tuneko Okazaki).
Because DNA polymerase III can add nucleotides only to the 3′ end of an existing strand, each Okazaki fragment is initiated by a
short RNA primer. Primase catalyzes the formation of the RNA primer for each upcoming Okazaki fragment as soon as the replication
fork has progressed a sufficient distance along the DNA. DNA polymerase III then adds nucleotides to the primer, generating an
Okazaki fragment that extends up to the 5′ end of the primer of the previously synthesized fragment.
3'
Finally, DNA polymerase I and other en- 5'
zymes replace the RNA primer of the pre- 3'
viously made Okazaki fragment with DNA, 5' 3'
and the enzyme DNA ligase covalently
joins successive Okazaki fragments into a 3' 5'
continuous strand of DNA.
5'
Ligase
199