Page 151 - Genetics_From_Genes_to_Genomes_6th_FULL_Part2
P. 151
310 Chapter 8 Gene Expression: The Flow of Information from DNA to RNA to Protein
17. You identify a proflavingenerated allele of a gene the electron microscope? Your figure should include
that produces a 110amino acid polypeptide rather hybridization involving both DNA strands (template
than the usual 157amino acid protein. After subjecting and RNAlike) as well as the mRNA.
this mutant allele to extensive proflavin mutagenesis, 23. In studying normal and mutant forms of a particular
you are able to find a number of intragenic suppressors human enzyme, a geneticist came across a particu
located in the part of the gene between the sequences larly interesting mutant form of the enzyme. The
encoding the N terminus of the protein and the original normal enzyme is 227 amino acids long, but the
mutation, but no suppressors located in the region mutant form was 312 amino acids long. The extra
between the original mutation and the sequences 85 amino acids occurred as a block in the middle
encoding the usual C terminus of the protein. Why of the normal sequence. The inserted amino acids
do you think this is the case? do not correspond in any way to the normal protein
18. Using recombinant DNA techniques (which will sequence. What are possible explanations for
be described in Chapter 9), it is possible to take the this phenomenon? How would you distinguish
DNA of a gene from any source and place it on a among them?
chromosome in the nucleus of a yeast cell. When 24. The Drosophila gene Dscam1 encodes proteins on the
you take the DNA for a human gene and put it into a surface of nerve cells (neurons) that govern neuronal
yeast cell chromosome, the altered yeast cell can connections. Each neuron has on its surface a single
make the human protein. But when you remove the Dscam1 protein of the tens of thousands that exist.
DNA for a gene normally present on yeast mitochon The particular Dscam1 protein a neuron expresses is
drial chromosomes and put it on a yeast chromosome in thought to tag the cell uniquely to determine the paths
the nucleus, the yeast cell cannot synthesize the correct of the axons and dendrites it will grow. Eukaryotic
protein, even though the gene comes from the same genes are monocistronic. How then can a single
organism. Explain. What would you need to do to ensure Dscam1 gene encode tens of thousands of different
that such a yeast cell could make the correct protein? proteins?
Section 8.2
Section 8.3
19. Describe the steps in transcription that require com
plementary base pairing. 25. Describe the steps in translation that require comple
20. Chapters 6 and 7 explained that mistakes made by mentary base pairing.
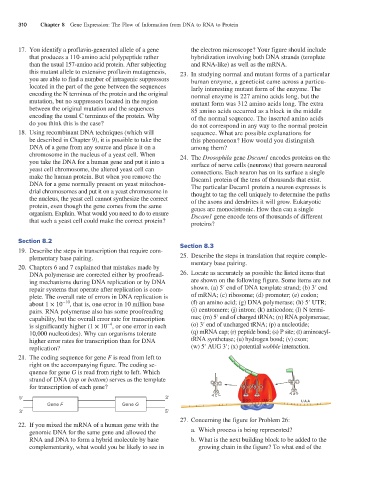
DNA polymerase are corrected either by proofread 26. Locate as accurately as possible the listed items that
ing mechanisms during DNA replication or by DNA are shown on the following figure. Some items are not
repair systems that operate after replication is com shown. (a) 5′ end of DNA template strand; (b) 3′ end
plete. The overall rate of errors in DNA replication is of mRNA; (c) ribosome; (d) promoter; (e) codon;
about 1 × 10 −10 , that is, one error in 10 million base (f) an amino acid; (g) DNA polymerase; (h) 5′ UTR;
pairs. RNA polymerase also has some proofreading (i) centromere; (j) intron; (k) anticodon; (l) N termi
capability, but the overall error rate for transcription nus; (m) 5′ end of charged tRNA; (n) RNA polymerase;
−4
is significantly higher (1 × 10 , or one error in each (o) 3′ end of uncharged tRNA; (p) a nucleotide;
10,000 nucleotides). Why can organisms tolerate (q) mRNA cap; (r) peptide bond; (s) P site; (t) aminoacyl
higher error rates for transcription than for DNA tRNA synthetase; (u) hydrogen bond; (v) exon;
replication? (w) 5′ AUG 3′; (x) potential wobble interaction.
21. The coding sequence for gene F is read from left to
right on the accompanying figure. The coding se
quence for gene G is read from right to left. Which
strand of DNA (top or bottom) serves as the template
for transcription of each gene?
A C C A U G
5' 3' GGG UA C UA A
Gene F Gene G
3' 5'
27. Concerning the figure for Problem 26:
22. If you mixed the mRNA of a human gene with the
genomic DNA for the same gene and allowed the a. Which process is being represented?
RNA and DNA to form a hybrid molecule by base b. What is the next building block to be added to the
complementarity, what would you be likely to see in growing chain in the figure? To what end of the