Page 65 - Genetics_From_Genes_to_Genomes_6th_FULL_Part1
P. 65
3.2 Extensions to Mendel for Two-Gene Inheritance 57
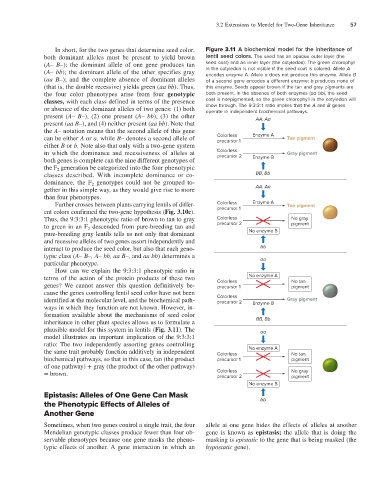
In short, for the two genes that determine seed color, Figure 3.11 A biochemical model for the inheritance of
both dominant alleles must be present to yield brown lentil seed colors. The seed has an opaque outer layer (the
(A– B–); the dominant allele of one gene produces tan seed coat) and an inner layer (the cotyledon). The green chlorophyll
(A– bb); the dominant allele of the other specifies gray in the cotyledon is not visible if the seed coat is colored. Allele A
encodes enzyme A. Allele a does not produce this enzyme. Allele B
(aa B–); and the complete absence of dominant alleles of a second gene encodes a different enzyme; b produces none of
(that is, the double recessive) yields green (aa bb). Thus, this enzyme. Seeds appear brown if the tan and gray pigments are
the four color phenotypes arise from four genotypic both present. In the absence of both enzymes (aa bb), the seed
classes, with each class defined in terms of the presence coat is nonpigmented, so the green chlorophyll in the cotyledon will
or absence of the dominant alleles of two genes: (1) both show through. The 9:3:3:1 ratio implies that the A and B genes
operate in independent biochemical pathways.
present (A– B–), (2) one present (A– bb), (3) the other AA, Aa
present (aa B–), and (4) neither present (aa bb). Note that
the A– notation means that the second allele of this gene
can be either A or a, while B– denotes a second allele of Colorless Enzyme A Tan pigment
precursor 1
either B or b. Note also that only with a two-gene system
in which the dominance and recessiveness of alleles at Colorless Gray pigment
precursor 2
both genes is complete can the nine different genotypes of Enzyme B
the F 2 generation be categorized into the four phenotypic
classes described. With incomplete dominance or co- BB, Bb
dominance, the F 2 genotypes could not be grouped to-
gether in this simple way, as they would give rise to more AA, Aa
than four phenotypes.
Further crosses between plants carrying lentils of differ- Colorless Enzyme A Tan pigment
ent colors confirmed the two-gene hypothesis (Fig. 3.10c). precursor 1
Thus, the 9:3:3:1 phenotypic ratio of brown to tan to gray Colorless No gray
to green in an F 2 descended from pure-breeding tan and precursor 2 pigment
pure-breeding gray lentils tells us not only that dominant No enzyme B
and recessive alleles of two genes assort independently and
interact to produce the seed color, but also that each geno- bb
typic class (A– B–, A– bb, aa B–, and aa bb) determines a aa
particular phenotype.
How can we explain the 9:3:3:1 phenotypic ratio in
terms of the action of the protein products of these two Colorless No enzyme A No tan
genes? We cannot answer this question definitively be- precursor 1 pigment
cause the genes controlling lentil seed color have not been
identified at the molecular level, and the biochemical path- Colorless Enzyme B Gray pigment
precursor 2
ways in which they function are not known. However, in-
formation available about the mechanisms of seed color
inheritance in other plant species allows us to formulate a BB, Bb
plausible model for this system in lentils (Fig. 3.11). The aa
model illustrates an important implication of the 9:3:3:1
ratio: The two independently assorting genes controlling
the same trait probably function additively in independent Colorless No enzyme A No tan
biochemical pathways, so that in this case, tan (the product precursor 1 pigment
of one pathway) + gray (the product of the other pathway)
= brown. Colorless No gray
precursor 2
pigment
No enzyme B
Epistasis: Alleles of One Gene Can Mask
the Phenotypic Effects of Alleles of bb
Another Gene
Sometimes, when two genes control a single trait, the four allele at one gene hides the effects of alleles at another
Mendelian genotypic classes produce fewer than four ob- gene is known as epistasis; the allele that is doing the
servable phenotypes because one gene masks the pheno- masking is epistatic to the gene that is being masked (the
typic effects of another. A gene interaction in which an hypostatic gene).