Page 120 - Genetics_From_Genes_to_Genomes_6th_FULL_Part2
P. 120
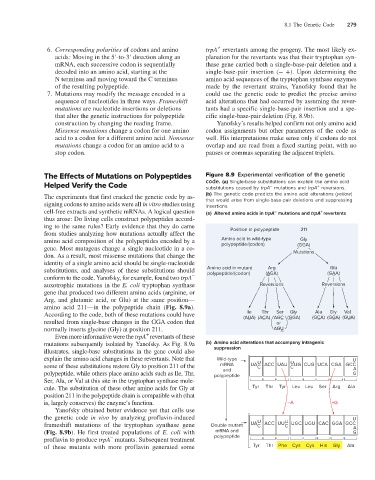
8.1 The Genetic Code 279
+
6. Corresponding polarities of codons and amino trpA revertants among the progeny. The most likely ex-
acids: Moving in the 5′-to-3′ direction along an planation for the revertants was that their tryptophan syn-
mRNA, each successive codon is sequentially thase gene carried both a single-base-pair deletion and a
decoded into an amino acid, starting at the single-base-pair insertion (− +). Upon determining the
N terminus and moving toward the C terminus amino acid sequences of the tryptophan synthase enzymes
of the resulting polypeptide. made by the revertant strains, Yanofsky found that he
7. Mutations may modify the message encoded in a could use the genetic code to predict the precise amino
sequence of nucleotides in three ways. Frameshift acid alterations that had occurred by assuming the rever-
mutations are nucleotide insertions or deletions tants had a specific single-base-pair insertion and a spe-
that alter the genetic instructions for polypeptide cific single-base-pair deletion (Fig. 8.9b).
construction by changing the reading frame. Yanofsky’s results helped confirm not only amino acid
Missense mutations change a codon for one amino codon assignments but other parameters of the code as
acid to a codon for a different amino acid. Nonsense well. His interpretations make sense only if codons do not
mutations change a codon for an amino acid to a overlap and are read from a fixed starting point, with no
stop codon. pauses or commas separating the adjacent triplets.
The Effects of Mutations on Polypeptides Figure 8.9 Experimental verification of the genetic
Helped Verify the Code code. (a) Single-base substitutions can explain the amino acid
+
−
substitutions caused by trpA mutations and trpA reversions.
The experiments that first cracked the genetic code by as- (b) The genetic code predicts the amino acid alterations (yellow)
signing codons to amino acids were all in vitro studies using that would arise from single-base-pair deletions and suppressing
insertions.
cell-free extracts and synthetic mRNAs. A logical question (a) Altered amino acids in trpA mutations and trpA revertants
–
+
thus arose: Do living cells construct polypeptides accord-
ing to the same rules? Early evidence that they do came
from studies analyzing how mutations actually affect the Position in polypeptide 211
amino acid composition of the polypeptides encoded by a Amino acid in wild-type Gly
gene. Most mutagens change a single nucleotide in a co- polypeptide/(codon) (GGA)
don. As a result, most missense mutations that change the Mutations
identity of a single amino acid should be single-nucleotide
Glu
Arg
substitutions, and analyses of these substitutions should Amino acid in mutant (AGA) (GAA)
polypeptide/(codon)
−
conform to the code. Yanofsky, for example, found two trpA – –
auxotrophic mutations in the E. coli tryptophan synthase Reversions Reversions
gene that produced two different amino acids (arginine, or
Arg, and glutamic acid, or Glu) at the same position—
amino acid 211—in the polypeptide chain (Fig. 8.9a).
Ala Gly Val
Ile Thr Ser Gly
According to the code, both of these mutations could have (AUA) (ACA) AGC (GGA) (GCA) (GGA) (GUA)
resulted from single-base changes in the GGA codon that – – – – – – –
or
normally inserts glycine (Gly) at position 211. AGU
–
+
Even more informative were the trpA revertants of these
mutations subsequently isolated by Yanofsky. As Fig. 8.9a (b) Amino acid alterations that accompany intragenic
illustrates, single-base substitutions in the gene could also suppression
explain the amino acid changes in these revertants. Note that Wild-type U
U
U
some of these substitutions restore Gly to position 211 of the mRNA UA ACC UAU UG CUG UCA CGA GCC A
C
C
and
polypeptide, while others place amino acids such as Ile, Thr, polypeptide G
Ser, Ala, or Val at this site in the tryptophan synthase mole-
cule. The substitution of these other amino acids for Gly at Tyr Thr Tyr Leu Leu Ser Arg Ala
position 211 in the polypeptide chain is compatible with (that
is, largely conserves) the enzyme’s function. –A +G
Yanofsky obtained better evidence yet that cells use
the genetic code in vivo by analyzing proflavin-induced U
U
U
frameshift mutations of the tryptophan synthase gene Double mutant UA ACC UU UGC UGU CAC GGA GCC A
C
C
(Fig. 8.9b). He first treated populations of E. coli with mRNA and G
−
proflavin to produce trpA mutants. Subsequent treatment polypeptide
of these mutants with more proflavin generated some Tyr Thr Phe Cys Cys His Gly Ala