Page 171 - Genetics_From_Genes_to_Genomes_6th_FULL_Part1
P. 171
5.5 Tetrad Analysis in Fungi 163
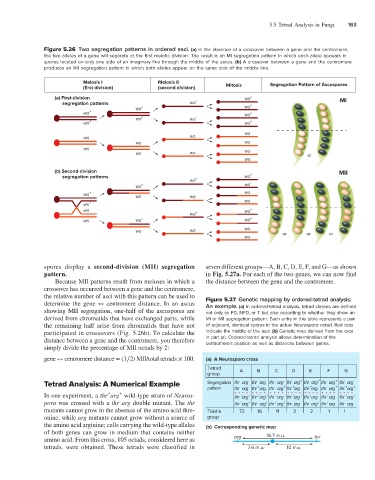
Figure 5.26 Two segregation patterns in ordered asci. (a) In the absence of a crossover between a gene and the centromere,
the two alleles of a gene will separate at the first meiotic division. The result is an MI segregation pattern in which each allele appears in
spores located on only one side of an imaginary line through the middle of the ascus. (b) A crossover between a gene and the centromere
produces an MII segregation pattern in which both alleles appear on the same side of the middle line.
Meiosis I Meiosis II Segregation Pattern of Ascospores
(first division) (second division) Mitosis
(a) First-division ws + MI
segregation patterns ws + +
ws + ws
ws + ws +
ws + ws +
ws + ws +
ws
ws ws
ws ws
ws ws
ws ws or
ws
(b) Second-division MII
segregation patterns + ws +
ws +
ws + ws
ws + ws ws ws
ws + ws
ws + ws +
ws
ws ws + ws +
ws ws ws or or or
ws
spores display a second-division (MII) segregation seven different groups—A, B, C, D, E, F, and G—as shown
pattern. in Fig. 5.27a. For each of the two genes, we can now find
Because MII patterns result from meioses in which a the distance between the gene and the centromere.
crossover has occurred between a gene and the centromere,
the relative number of asci with this pattern can be used to Figure 5.27 Genetic mapping by ordered-tetrad analysis:
determine the gene ↔ centromere distance. In an ascus An example. (a) In ordered-tetrad analysis, tetrad classes are defined
showing MII segregation, one-half of the ascospores are not only as PD, NPD, or T but also according to whether they show an
derived from chromatids that have exchanged parts, while MI or MII segregation pattern. Each entry in this table represents a pair
the remaining half arise from chromatids that have not of adjacent, identical spores in the actual Neurospora octad. Red dots
participated in crossovers (Fig. 5.26b). To calculate the indicate the middle of the asci. (b) Genetic map derived from the data
distance between a gene and the centromere, you therefore in part (a). Ordered-tetrad analysis allows determination of the
centromere’s position as well as distances between genes.
simply divide the percentage of MII octads by 2:
gene ↔ centromere distance = (1∕2) MII/total tetrads × 100. (a) A Neurospora cross
Tetrad A B C D E F G
group
+
+
Tetrad Analysis: A Numerical Example Segregation thr arg thr arg thr arg thr arg + thr arg thr arg thr arg
+
pattern thr arg thr arg thr arg thr + arg thr arg thr arg thr + arg +
+
+
+
+
+
In one experiment, a thr arg wild-type strain of Neuros- thr arg + thr arg + thr arg thr + arg + thr arg thr arg thr + arg +
+
+
+
+
+
pora was crossed with a thr arg double mutant. The thr thr arg + thr arg + thr arg thr arg thr arg thr arg thr arg
+
+
+
+
+
mutants cannot grow in the absence of the amino acid thre- Total in 72 16 11 2 2 1 1
onine, while arg mutants cannot grow without a source of group
the amino acid arginine; cells carrying the wild-type alleles (b) Corresponding genetic map
of both genes can grow in medium that contains neither
amino acid. From this cross, 105 octads, considered here as arg 16.7 m.u. thr
tetrads, were obtained. These tetrads were classified in 7.6 m.u. 10 m.u.